Kinesin-4 KIF21B is a potent microtubule pausing factor
Figures
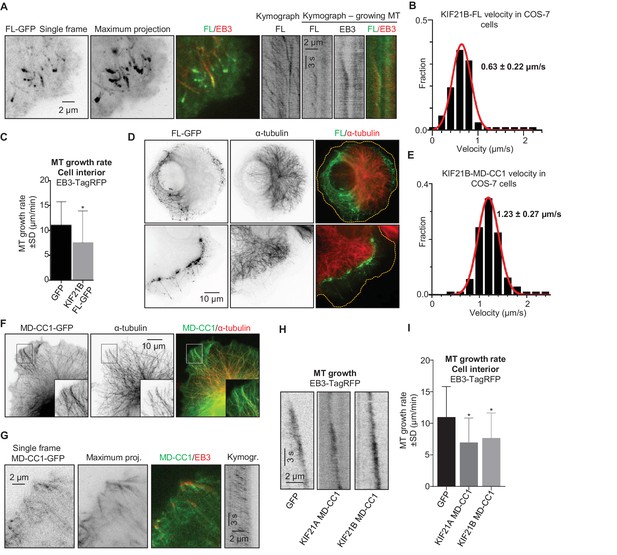
KIF21B inhibits MT growth in cells.
(A) COS-7 cells were transiently transfected with KIF21B-FL-GFP and EB3-TagRFP-T and imaged using TIRF microscopy. Represented are a single-frame, maximum intensity projection of 500 frames for the GFP channel and an overlay of a single GFP frame in green and TagRFP-T in red. Kymographs illustrate the motility of KIF21B along the MT and its significant accumulation at a stationary but not a growing MT plus end. (B) Histogram of KIF21B-FL-GFP kinesin velocities in COS-7 cells is shown with black bars. Red line shows fitting with a normal distribution. n = 378 in 10 cells in two independent experiments. (C) Quantification of MT growth rate, measured by tracking EB3 labeled comets in cell interior. Three to ten MTs per cell were analyzed; n = 183 in 21 cells for GFP control, n = 214 in 12 cells for KIF21B-FL-GFP expressing cells, two independent experiments, p<0.0001, Mann-Whitney U test (indicated by an asterisk). (D) COS-7 cells were transiently transfected with KIF21B-FL-GFP, fixed the next day and stained for α-tubulin. Cell edges are indicated with yellow dashed lines in the overlay. (E) Histogram of KIF21B-MD-CC1-GFP velocities in COS-7 cells is shown with black bars. Red line shows fitting with a normal distribution. n = 431 in 14 cells in two independent experiments. (F) COS-7 cells transiently transfected with KIF21B-MD-CC1-GFP were fixed and stained for α-tubulin. (G) COS-7 cells were transiently transfected with KIF21B-MD-CC1-GFP and EB3-TagRFP-T and imaged using TIRF microscopy. Represented are a single-frame, maximum intensity projection of 500 frames for the GFP channel, an overlay of a single GFP frame in green and TagRFP-T in red and a kymograph along one of the EB3-labeled MTs showing the motility of the kinesin along the MT. (H) Kymographs showing EB3-TagRFP-T comet displacement in control COS-7 cells or cells expressing the MD-CC1 fragments of KIF21A or KIF21B. (I) Quantification of MT growth rate illustrated in H. n = 183 in 21 cells for GFP control, n = 136 in 15 cells for KIF21A-MD-CC1-GFP, n = 179 in 22 cells for KIF21B-MD-CC1-GFP, two independent experiments, p<0.0001, Mann-Whitney U test (indicated by asterisks).
-
Figure 1—source data 1
An excel sheet with numerical data on the quantification of kinesin velocities and MT growth rate in COS-7 cells represented as plots in Figure 1B,C,E,I.
- https://doi.org/10.7554/eLife.24746.004
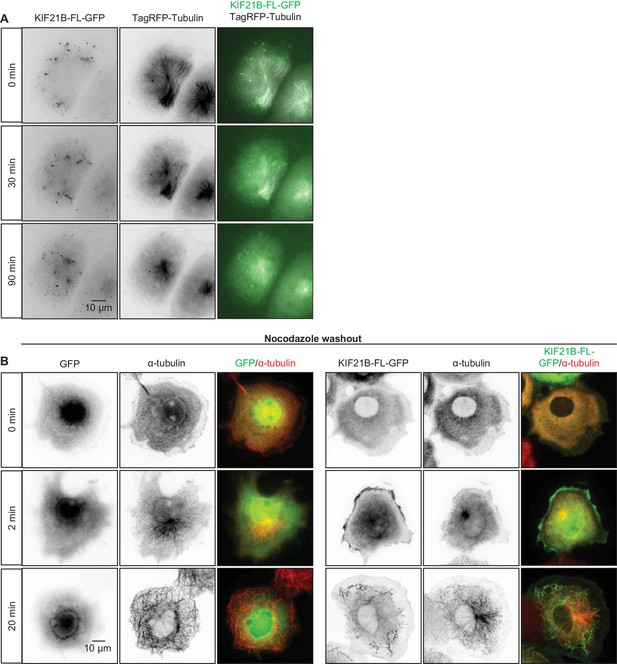
Effects of KIF21B expression on MT organization and regrowth in cells
(A) Time-lapse imaging of transiently transfected COS-7 cells expressing KIF21B-FL-GFP and TagRFP-tubulin. Yellow dashed lines in the overlay indicate the cell edge. (B) Nocodazole washout experiments of COS-7 cells expressing GFP or KIF21B-FL-GFP. Cells were transiently transfected with the indicated proteins and treated with 5 μM nocodazole for 2 hr. Subsequently, nocodazole was washed out and cells were fixed at the indicated time points. Antibodies against α-tubulin were used for cell staining. Yellow dashed lines in the overlay indicate the cell edge.
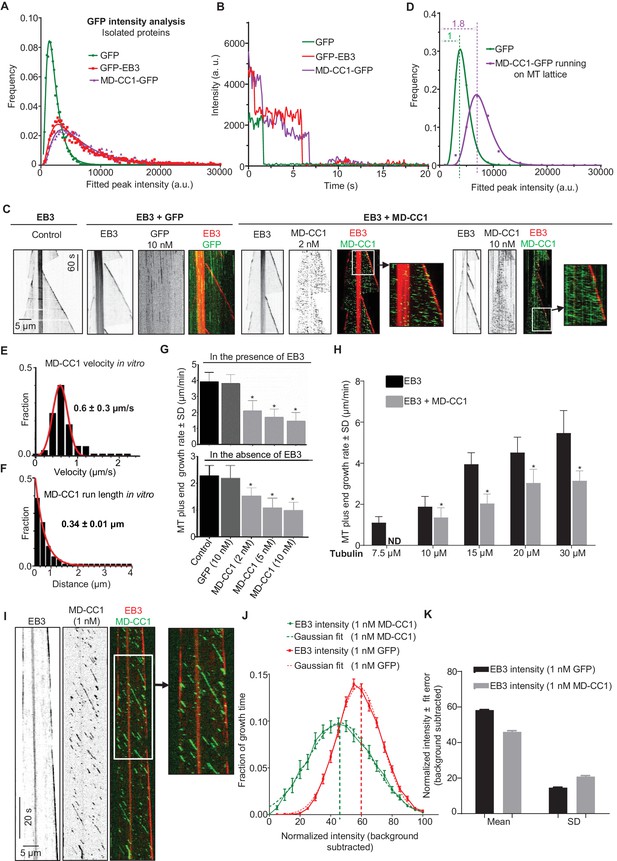
Dimeric motor domain of KIF21B slows down MT polymerization in vitro.
(A) Histograms of fluorescence intensities at the initial moment of observation of single molecules of the indicated proteins immobilized on coverslips (symbols) and the corresponding fits with lognormal distributions (lines). n = 3107, 5802 and 4674 molecules and fluorophore density was 0.15, 0.28 and 0.23 µm−2 for GFP, GFP-EB3 and KIF21B-MD-CC1-GFP proteins. (B) Representative photobleaching time traces of GFP, GFP-EB3 and KIF21B-MD-CC1-GFP individual molecules (background subtracted). (C) Kymographs illustrating the dynamics of MTs grown in vitro in the presence of 20 nM mCherry-EB3 alone, with 10 nM purified GFP or with 2 and 10 nM KIF21B-MD-CC1-GFP. Zooms of the boxed areas are shown on the right. Kymographs were generated from movies acquired using a Photometrics Evolve 512 EMCCD camera (Roper Scientific) (stream acquisition, exposure time 500 ms). (D) Histograms of fluorescence intensities of single GFP molecules immobilized on coverslips and KIF21B-MD-CC1-GFP moving on MTs in a separate chamber on the same coverslip (symbols) and the corresponding fits with lognormal distributions (lines). n = 4815 and 1381 molecules; fluorophore density was 0.16 and 0.09 µm−2 for GFP and KIF21B-MD-CC1-GFP proteins (for the latter, MT-containing regions were manually selected for analysis). Dashed lines show corresponding relative median values. (E) Histogram of KIF21B-MD-CC1-GFP velocities in vitro is shown with black bars. Red line shows fitting with a normal distribution. n = 675 in two independent experiments. (F) Histogram of KIF21B-MD-CC1-GFP run lengths in vitro is shown with black bars. Red line shows fitting with an exponential distribution. n = 675 in two independent experiments. (G) Upper panel - quantification of the MT growth rate illustrated in C. n = 71 for control, n = 65 for purified GFP, n = 71, 67 and 54 for 2, 5 and 10 nM KIF21B-MD-CC1-GFP, respectively. Lower panel shows quantification of the MT growth rate with 15 µM tubulin alone or with 10 nM purified GFP or with 2, 5 and 10 nM KIF21B-MD-CC1-GFP as illustrated in Figure 2—figure supplement 2. n = 67 for control, n = 57 for purified GFP, n = 71, 66 and 80 for 2, 5 and 10 nM KIF21B-MD-CC1-GFP, respectively, two independent experiments, p<0.0001, Mann-Whitney U test (indicated by asterisks). (H) Quantification of the MT growth rate with different concentrations of tubulin along with 20 nM EB3 in the absence and presence of 2 nM KIF21B-MD-CC1-GFP as illustrated in Figure 2—figure supplement 3. ND; not determined, n = 71 for all conditions. two independent experiments, p<0.0001, Mann-Whitney U test (indicated by asterisks). (I) Kymographs illustrating the dynamics of MTs grown in vitro in the presence of 20 nM EB3 and 1 nM KIF21B-MD-CC1-GFP. Zoom of the boxed area is shown on the right. Kymographs were generated from a movie acquired using Photometrics Evolve 512 EMCCD camera (Roper Scientific) (stream acquisition, exposure time 100 ms). (J) Distribution of EB3 fluorescence intensity fluctuations over time (normalized to its maximum value during a course of a growth event) at MT tip in the presence of 1 nM GFP or 1 nM KIF21B-MD-CC1-GFP (solid line) with Gaussian fit (dotted line). n = 25 in both cases. Thick dotted lines show the peak of the Gaussian fitting. MT dynamics assay was performed in the presence of 15 µM tubulin, 20 nM EB3 and 1 nM GFP or 1 nM KIF21B-MD-CC1-GFP in the separate chambers of the same coverslip. (I) Plot of the mean and SD values of Gaussian fits shown in Figure 2J.
-
Figure 2—source data 1
An excel sheet with numerical data on the quantification of KIF21B-MD-CC1-GFP dimer analysis, photobleaching-step analysis, velocities, run length, effects on MT growth rate and distribution of EB3 fluorescence intensity represented as plots in Figures 2A,B,D,E–H,J.
- https://doi.org/10.7554/eLife.24746.007
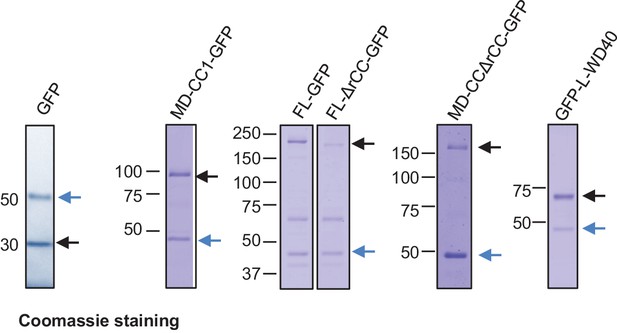
Coomassie blue stained gels with purified GFP, KIF21B-FL-GFP and its deletion mutants.
Protein purification was performed using TEV protease cleavage as described in the Materials and Methods section. Black arrows indicate isolated proteins; blue arrows indicate the TEV protease.
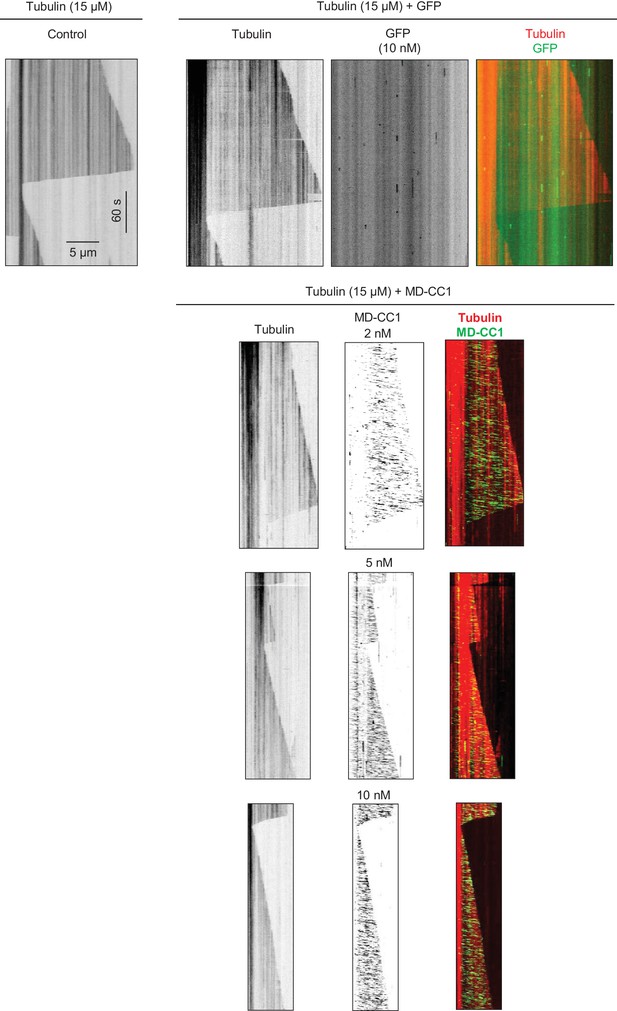
Kymographs illustrating in vitro dynamics of MTs grown in the presence of 15 µM tubulin in the absence and presence of 10 nM purified GFP or 2, 5 and 10 nM KIF21B-MD-CC1-GFP.
Kymographs were generated from the movies of 600 frames (stream acquisition, exposure time 500 ms) using Photometrics Evolve 512 EMCCD camera (Roper Scientific).
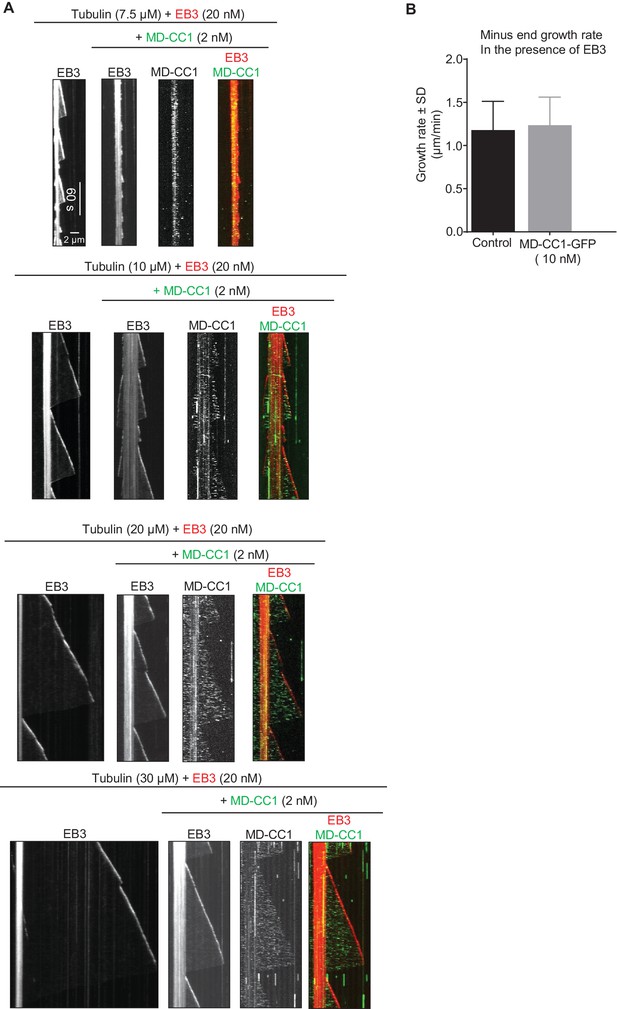
Effects of the dimeric motor domain of KIF21B on MT polymerization in vitro
(A) Kymographs illustrating the in vitro dynamics of MTs grown in the presence of different concentrations of tubulin along with 20 nM EB3 in the absence and presence of 2 nM KIF21B-MD-CC1-GFP. Kymographs were generated from the movies of 600 frames (stream acquisition, exposure time 500 ms) using Photometrics Evolve 512 EMCCD camera (Roper Scientific). (B) Quantification of the velocity of MT minus end growth in the presence of 15 µM tubulin and 20 nM mCherry-EB3 without (control) or with 10 nM KIF21B-MD-CC1-GFP. n = 50 in both cases.
-
Figure 2—Figure Supplement 3—Source Data 1
An excel sheet with numerical data on the quantification of the MT minus end growth rates represented as plot in Figure 2—figure supplement 3B.
- https://doi.org/10.7554/eLife.24746.011
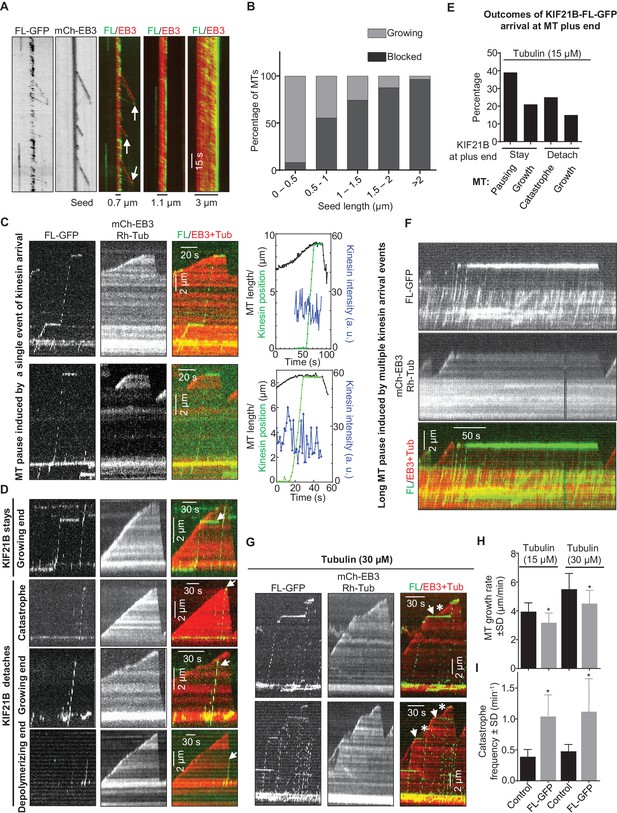
KIF21B can induce MT pausing or catastrophe in vitro.
(A) Kymographs showing the behavior of KIF21B in in vitro reconstitution assays on dynamic MTs grown from Rhodamine-tubulin-labeled seeds in the presence of 15 µM tubulin, 100 nM mCherry-EB3 (red) and 3 nM KIF21B-FL-GFP (green). Kymographs were generated from movies acquired using a Photometrics Evolve 512 EMCCD camera (Roper Scientific) (stream acquisition, exposure time 500 ms). Pausing and catastrophe events are indicated by arrows. (B) Quantification of MT seed length-dependent blocking of MT growth by 0.5 nM KIF21B- FL-GFP in the presence of 20 nM mCherry-EB3. 188 MT seeds of different lengths were analyzed in four independent experiments. (C) Kymographs illustrating pausing events induced by KIF21B-FL-GFP (0.5 nM) on dynamic MTs in vitro in the presence of 15 µM tubulin, 20 nM mCherry-EB3, 3% Rhodamine-tubulin. MTs were grown from GMPCPP-stabilized seeds labeled with Rhodamine-tubulin. Kymographs were generated from the movies acquired using a CoolSNAP HQ2 CCD camera (Roper Scientific) with a 1.2-s interval between frames and an exposure time of 100 ms. The rightmost panels show tracked positions of the kinesins and MT tips together with the fluorescence intensity of the kinesins over time for the corresponding kymographs. (D) Kymographs illustrating various events induced by KIF21B-FL-GFP (0.5 nM) on dynamic MTs in vitro in the presence of 15 µM tubulin, 20 nM mCherry-EB3 and 3% Rhodamine-tubulin. MTs were grown from GMPCPP-stabilized seeds labeled with Rhodamine-tubulin. Different events are indicated by arrows. Kymographs were generated from movies acquired as described for Figure 3C. (E) Quantification of different events observed after KIF21B-FL-GFP (0.5 nM) reaches a growing MT plus end, as illustrated in C and D. n = 132 events, four independent experiments were analyzed. (F) Kymograph illustrating a long pause event induced by multiple KIF21B-FL-GFP molecules on dynamic MTs in vitro in the presence of 15 µM tubulin, 20 nM mCherry-EB3 and 3% Rhodamine-tubulin in solution. Kymographs are generated from a movie acquired as described for Figure 3C. (G) Kymographs illustrating the effects of KIF21B-FL-GFP (0.5 nM) on dynamic MTs in vitro in the presence of 30 µM tubulin with 3% Rhodamine-tubulin and 20 nM mCherry-EB3. MTs were grown from GMPCPP-stabilized seeds labeled with Rhodamine-tubulin. The arrows show the position of KIF21B at the site of MT pause and the asterisk indicates the growing MT tip beyond the position of KIF21B binding; note that the slope of the kymograph after KIF21B attachment is less steep than before, indicating that the growth rate is reduced. Kymographs are generated from movies acquired as described for Figure 3C. (H, I) Quantification of MT growth rate and catastrophe frequency in vitro in the presence of 15 or 30 µM tubulin with 20 nM mCherry-EB3 alone or together with 0.5 nM KIF21B-FL-GFP. MTs were grown in the presence of 3% Rhodamine-tubulin. For 15 µM tubulin, n = 71 for control and n = 100 for KIF21B-FL-GFP, three independent experiments. For 30 µM tubulin, n = 71 for control and n = 80 for KIF21B-FL-GFP, three independent experiments, p<0.0001 Mann-Whitney U test (indicated by asterisks).
-
Figure 3—source data 1
An excel sheet with numerical data on the quantification of KIF21B-FL seed blocking activity, pause induction, effects on MT growth rate and catastrophe frequency and outcomes of KIF21B-FL-GFP arrival at MT plus ends represented as plots in Figure 3B,C,E,H,I.
- https://doi.org/10.7554/eLife.24746.013
Characterization of full-length KIF21B in vitro
(A) Histograms of fluorescence intensities at the initial moment of observation of single molecules of the indicated proteins immobilized on coverslips (symbols) and the corresponding fits with lognormal distributions (lines). n = 5063, 8830 and 12230 molecules; fluorophore density was 0.23, 0.4 and 0.45 µm−2 for GFP, GFP-EB3 and KIF21B-FL-GFP proteins. (B) Representative photobleaching time traces of GFP, EB3-GFP and KIF21B-FL-GFP individual molecules (background subtracted). (C) Kymographs illustrating MT dynamics in vitro in the presence of 15 µM tubulin with 3% Rhodamine-tubulin and 0.5 nM KIF21B-FL-GFP. Kymographs were generated from the movies acquired using CoolSNAP HQ2 CCD camera (Roper Scientific) with a 1.2-s interval between frames and an exposure time of 100 ms.
-
Figure 3—figure supplements 1—source data 1
An excel sheet with numerical data on the quantification of the KIF21B-FL dimer and photobleaching step analysis represented as plots in Figure 3—figure supplement 1A,B.
- https://doi.org/10.7554/eLife.24746.015
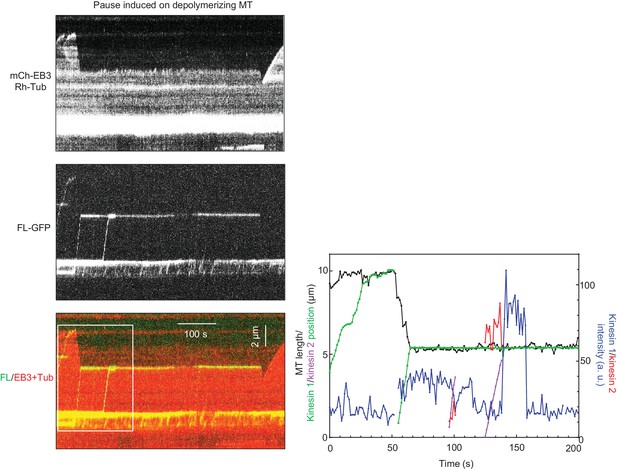
KIF21B-FL-GFP induces pausing of a depolymerizing MT.
The rightmost panel shows tracked positions of the kinesins and the MT tip together with the fluorescence intensities of the kinesins over time (white boxed area in kymograph). See also Supplemental Video 1. Kymograph was generated from a movies acquired using CoolSNAP HQ2 CCD camera (Roper Scientific) with a 1.2-s interval between frames and an exposure time of 100 ms.
-
Figure 3—Figure Supplement 2—Source Data 1
An excel sheet with numerical data on the quantification of tracked positions of the kinesins and the MT tip together with the fluorescence intensities of the kinesins over time represented as plot in Figure 3—figure supplement 2.
- https://doi.org/10.7554/eLife.24746.017
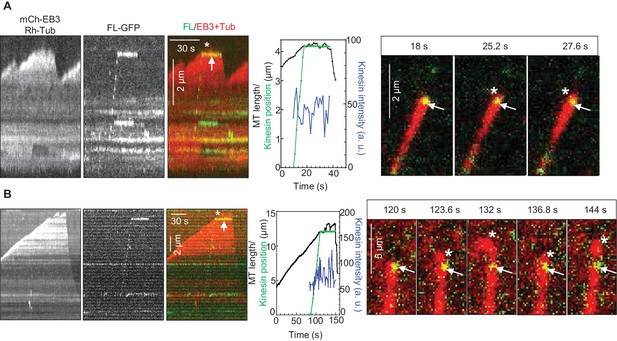
KIF21B molecules persisting on a MT tip can perturb MT growth.
(A, B) Kymographs illustrating perturbation of MT growth in vitro by 0.5 nM KIF21B-FL-GFP in the presence of 15 µM tubulin with 3% Rhodamine-tubulin and 20 nM mCherry-EB3. Kymographs were generated from movies acquired as described for Figure 3C. Positions of the kinesins and MT tips together with the fluorescence intensity of the kinesins over time for the corresponding kymographs are also illustrated. Time lapse images on the right of the kymographs illustrate short excursions of MT plus tip (A) or curling of MT plus tip (B) after the binding of KIF21B-FL-GFP to the MT plus end. The position of the kinesin on the MT is indicated by arrows. Asterisks show the position of growing MT tips extending beyond the point of KIF21B attachment. See also Supplemental Video 2.
-
Figure 4—source data 1
An excel sheet with numerical data on the quantification of tracking of kinesins and MT tips over time represented as plots in Figure 4A,B.
- https://doi.org/10.7554/eLife.24746.020
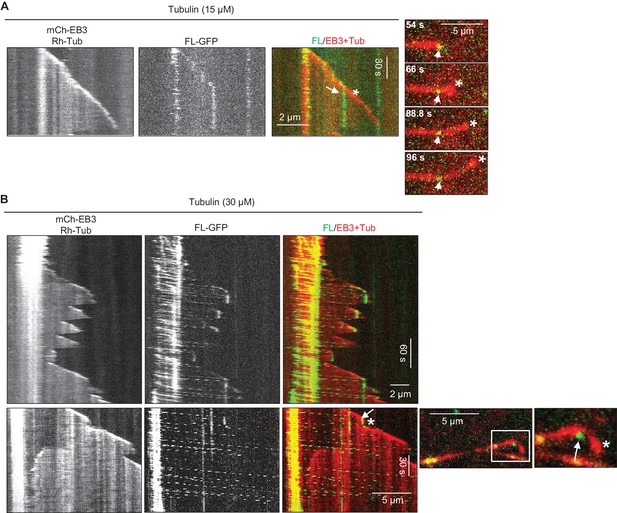
Perturbation of MT growth in vitro by full-length KIF21B
(A, B) Kymographs illustrating perturbation of MT growth in vitro by 0.5 nM KIF21B-FL-GFP in the presence of 15 and 30 µM tubulin with 3% Rhodamine-tubulin and 20 nM mCherry-EB3. Time lapse images on the right illustrate MT plus tip curling after the binding of KIF21B-FL-GFP to the MT plus end. The position of the kinesin on the MT is indicated by arrows. Asterisks show the position of growing MT tips extending beyond the point of KIF21B attachment. Boxed area is zoomed. See also Supplemental Video 2. Kymographs were generated from movies acquired using CoolSNAP HQ2 CCD camera (Roper Scientific) with a 1.2-s interval between frames and an exposure time of 100 ms.
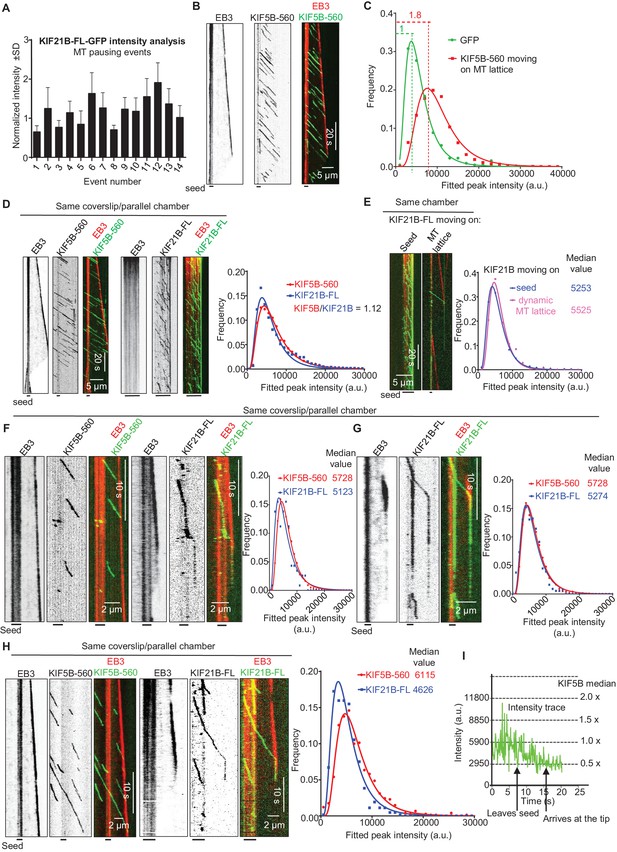
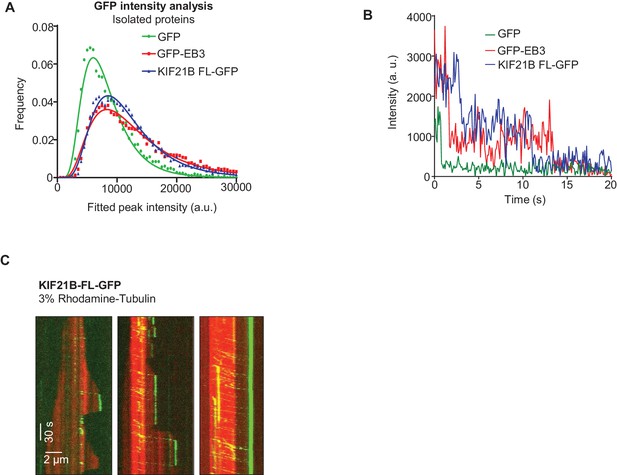
Single events of kinesin arrival to the MT plus end can induce MT pausing.
(A) GFP intensity analysis of kinesins during MT pausing events. Values are normalized to the GFP intensity of proteins immobilized on the same coverslip in areas devoid of MTs. Data are from two independent experiments. (B) Kymographs showing the behavior of KIF5B-560-GFP in an in vitro reconstitution assay on dynamic MTs grown from Rhodamine-tubulin-labeled seeds in the presence of 15 µM tubulin, 20 nM mCherry-EB3 (red) and 5 nM KIF5B-560-GFP (green). (C) Histograms of fluorescence intensities of single GFP molecules immobilized on coverslips (initial moment of observation of single molecules) or KIF5B-560-GFP moving on MTs in a separate chamber on the same coverslip (symbols) and the corresponding fits with lognormal distributions (lines). n = 452 and 2040 molecules; fluorophore density was 0.05 and 0.09 µm−2 for GFP and KIF5B-560-GFP proteins (for the latter, MT-containing regions were manually selected for analysis). Dashed lines show the corresponding relative median values. (D) Kymographs showing the behavior of 5 nM KIF5B-560-GFP (moving on dynamic MTs, green) and 0.5 nM KIF21B-FL-GFP (moving on seeds, green) in an in vitro reconstitution assay with MTs grown from Rhodamine-tubulin-labeled seeds in the presence of 15 µM tubulin and 20 nM mCherry-EB3 (red). Histograms illustrate fluorescence intensities of KIF5B-560-GFP moving on MTs and KIF21B-FL-GFP moving on seeds in separate chambers on the same coverslip (symbols) and the corresponding fits with lognormal distributions (lines). n = 5123 and 8728 molecules; fluorophore density was 0.15 and 0.18 µm−2 for KIF5B-560-GFP and KIF21B-FL-GFP proteins (MT-containing regions were manually selected for analysis). Ratio of the corresponding median values is also indicated. (E) Kymographs illustrating KIF21B-FL-GFP (0.5 nM) movement on seeds and dynamic MTs, histograms of the corresponding fluorescence intensities measured within the same sample (symbols) and their fits with lognormal distributions (lines). Median values are also indicated. (F–H) Kymographs illustrating motility of KIF5B-560-GFP (5 nM) on dynamic MTs and KIF21B-FL-GFP (0.5 nM) moving from an MT seed to the dynamic MT lattice and inducing a MT pause in an in vitro reconstitution assay with MTs grown from Rhodamine-tubulin-labeled seeds in the presence of 15 µM tubulin and 20 nM mCherry-EB3 (red). Histograms show fluorescence intensities of motile KIF5B-560-GFP molecules and KIF21B-FL-GFP inducing a MT pause in separate chambers on the same coverslip (symbols) and the corresponding fits with lognormal distributions (lines). Median values are also indicated. In all the conditions, kymographs were generated from movies of 1500 frames (stream acquisition, exposure time 100 ms) using Photometrics Evolve 512 EMCCD camera (Roper Scientific). Positions of seeds in each kymograph are indicated. (I) Fitted peak intensity time trace for the trajectory of a moving KIF21B-FL-GFP molecule from the event shown in Figure 5H. Dashed lines correspond to the scaled values of median fluorescence fitted peak intensity of KIF5B-560-GFP molecules moving on dynamic MT in a parallel chamber on the same coverslip.
-
Figure 5—source data 1
An excel sheet with numerical data on the quantification of KIF21B-FL intensity during MT pausing events, KIF5B-560 dimer analysis and comparison of fluorescence intensities of KIF5B-560 with KIF21B-FL represented as plots in Figure 5A,C,D–I.
- https://doi.org/10.7554/eLife.24746.024
-
Figure 5—figure Supplement 2—Source data 1
An excel sheet with numerical data on the quantification of photobleaching traces of KIF21B-FL-GFP represented as plots in Figure 5—figure supplement 2.
- https://doi.org/10.7554/eLife.24746.025
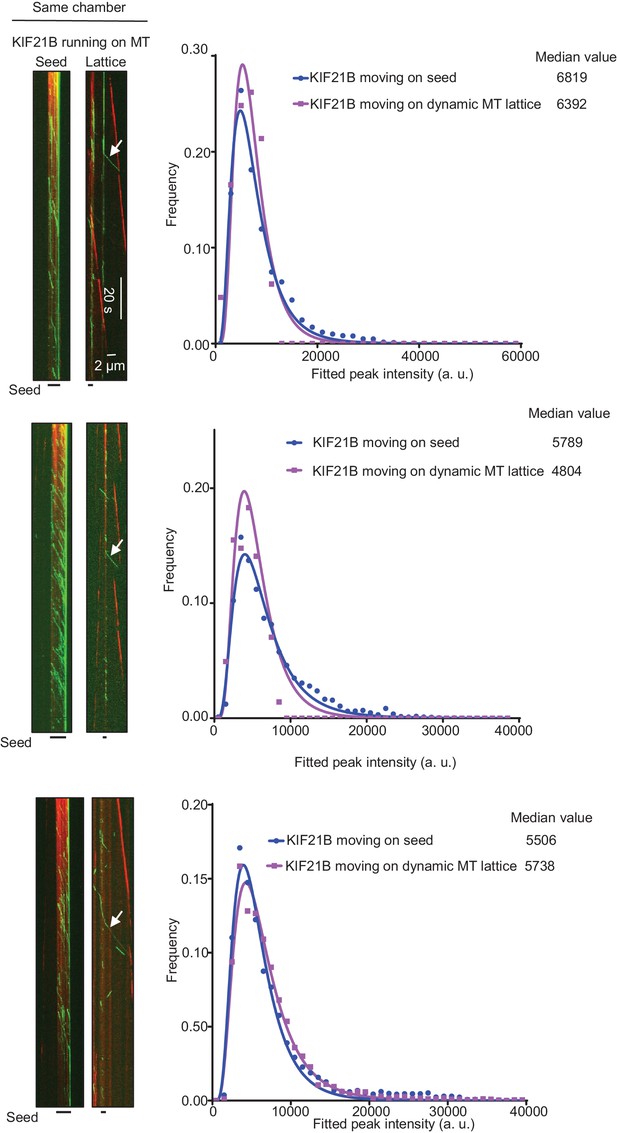
Kymographs illustrating KIF21B-FL-GFP (0.5 nM) moving on seeds and dynamic MTs in in vitro reconstitution assays, histograms of the corresponding fluorescence intensities (symbols) and the corresponding fits with lognormal distributions (lines).
Median values are also indicated.
-
Figure 5—figure supplements 1—Source data 1
An excel sheet with numerical data on the quantification of KIF21B-FL fluorescence intensities represented as plots in Figure 5—figure supplement 1.
- https://doi.org/10.7554/eLife.24746.027
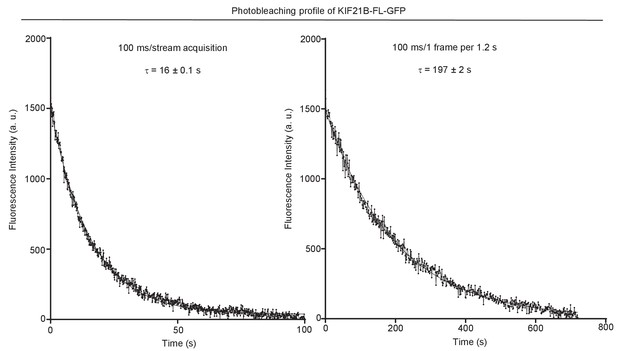
Characteristic photobleaching traces of KIF21B-FL-GFP under two different imaging conditions.
KIF21B-FL-GFP immobilized on coverslip was exposed to low laser power (used for imaging shown in Figures 3–5 and 7) with 100 ms/stream acquisition or 100 ms exposure time, 1 frame per 1.2 s. Curves were fitted with one-phase exponential decay.
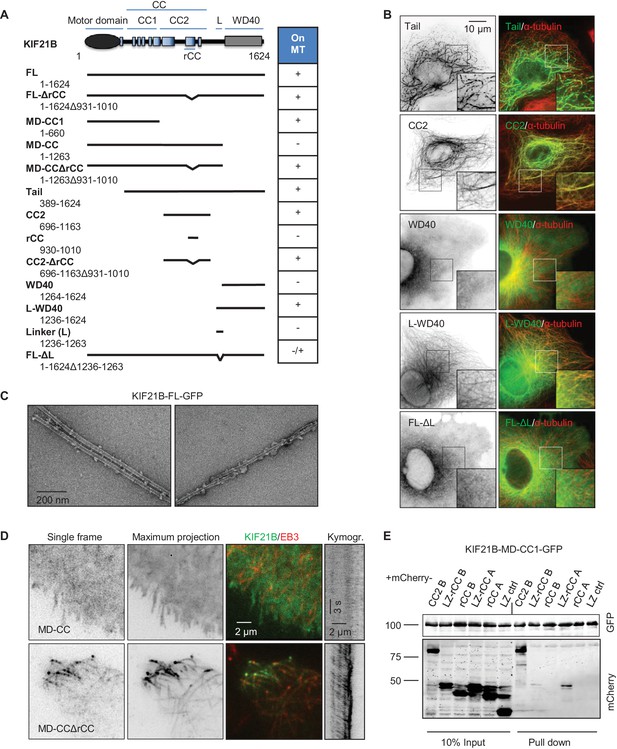
Mapping of the MT-binding domains in the tail of KIF21B.
(A) Overview of deletion mutants used in this study. Colocalization of the GFP-tagged KIF21B deletion mutants with MTs in transiently transfected COS-7 cells is indicated. +, localization to MTs, -, diffuse distribution, -/+, diffuse in most cells, with occasional MT localization observed in some cells. (B) COS-7 cells were fixed one day after transient transfection with the indicated constructs and stained for α-tubulin. (C) Electron micrographs of negatively stained taxol-stabilized MTs in complex with KIF21B-FL-GFP. (D) Live imaging of COS-7 cells transiently transfected with KIF21B-MD-CC-GFP or MD-CCΔrCC-GFP and EB3-TagRFP-T. Represented are a single-frame, maximum intensity projection of 500 frames for the GFP channel, an overlay of single GFP frame in green and TagRFP-T in red and a kymograph along one of the EB3-labeled MTs showing kinesin motility. (E) Streptavidin pull down assay with the extracts of HEK293T cell expressing BirA, KIF21B-MD-CC1-GFP-TEV-Bio and the indicated mCherry-labeled proteins. A and B stand for KIF21A and KIF21B; LZ, leucine zipper from GCN4 used for dimerization. The other abbreviations are explained in panel A. The results were analyzed by Western blotting with the antibodies against the GFP- and mCherry.
-
Figure 6—figure supplement 4—source data 1
An excel sheet with numerical data on the quantification of far-UV CD spectra (inset) and thermal unfolding profile of recombinant KIF21B rCC1 represented as plots in Figure 6—figure supplement 4A.
- https://doi.org/10.7554/eLife.24746.030
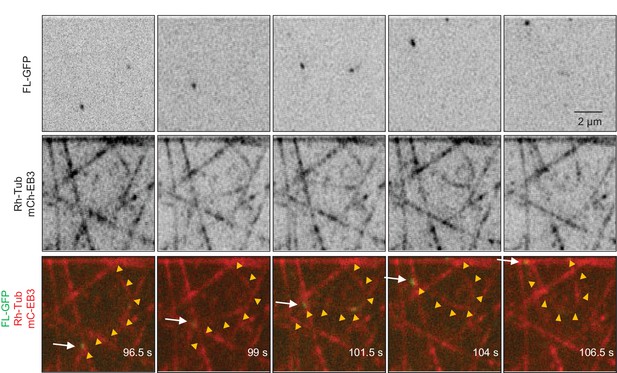
In vitro reconstitution of MT growth in the presence of 20 nM mCherry-EB3, 3% Rhodamine-tubulin and 0.5 nM KIF21B-FL-GFP.
KIF21B-FL-GFP is attached to one MT and walks along another one, causing MT bending. Arrow indicates position of KIF21B-FL-GFP, yellow arrowheads trace the MT that bends.
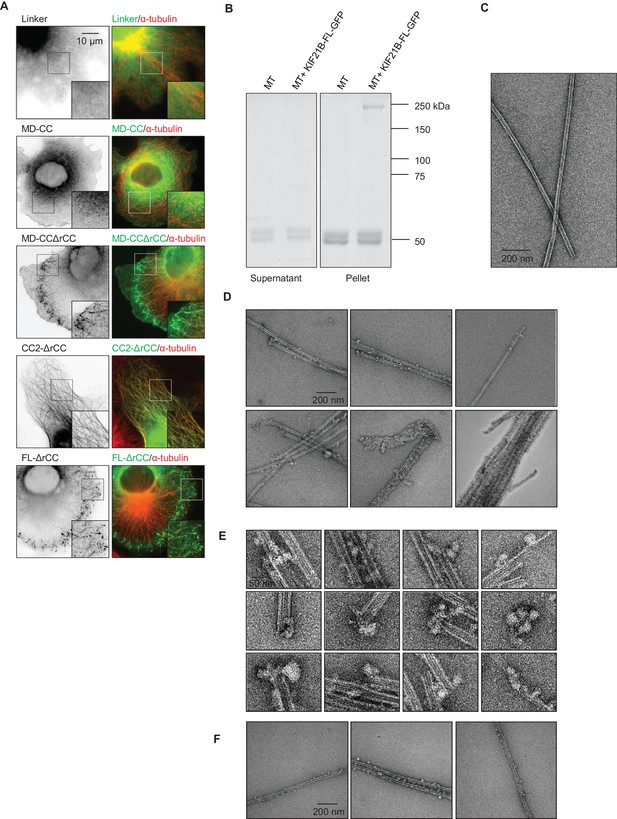
(A) COS-7 cells transiently transfected with the indicated KIF21B-GFP deletion constructs and stained for α-tubulin. (B) MT pelleting assay of taxol-stabilized MTs incubated with KIF21B-FL-GFP. Coomassie-stained SDS-PAGE of supernatant and pellet fractions of MTs alone or MTs incubated with KIF21B-FL-GFP are shown. (C) Electron micrograph of a negatively stained taxol-stabilized MT control specimens. (D) Gallery of electron micrographs of negatively stained taxol-stabilized MT specimens obtained in the presence of KIF21B-FL-GFP. (E) High-magnification views of taxol-stabilized MT ends decorated with KIF21B-FL-GFP. (F) Gallery of electron micrographs of negatively stained GMPCPP-MT specimens obtained in the presence of KIF21B-FL-GFP.

Alignment of human KIF21A and KIF21B sequences.
Different protein domains and the autoinhibitory region in the coiled coil domain described for KIF21A (van der Vaart et al., 2013) are indicated, and CFEOM1-associated mutations found in KIF21A are boxed. The KIF21B sequence shown here corresponds to the longest KIF21B isoform (Accession number O75037); a shorter isoform (Accession number BAA32294), which misses the amino acids 1269–1281 (underlined), was used in this study.
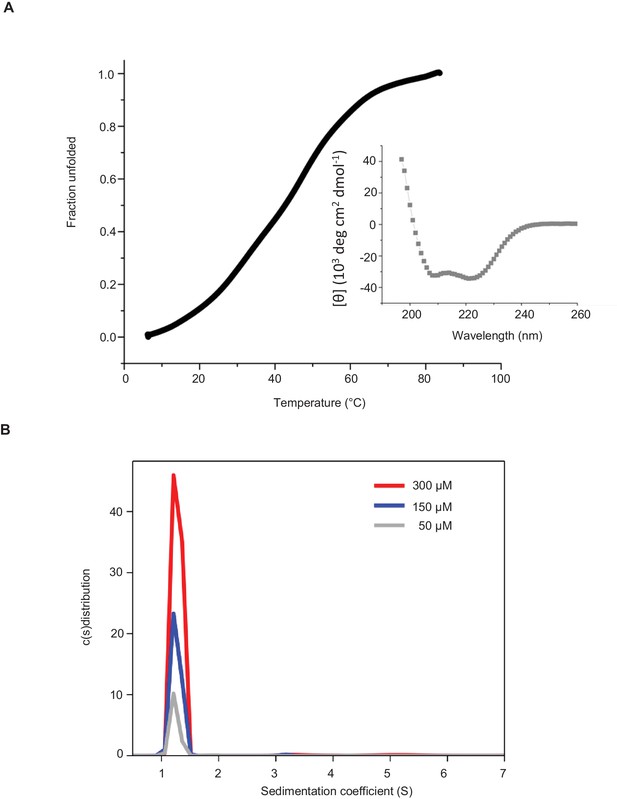
(A) Far-UV CD spectra (inset) and thermal unfolding profile of recombinant KIF21B rCC. CD measurements, performed in PBS at a protein concentration of 0.166 mg/ml. (B) Oligomerization state of recombinant KIF21B rCC determined by sedimentation velocity AUC at 20°C and at three different protein concentrations.
The WD40 domain and the autoinhibitory coiled coil region contribute to the pause-promoting activity of KIF21B.
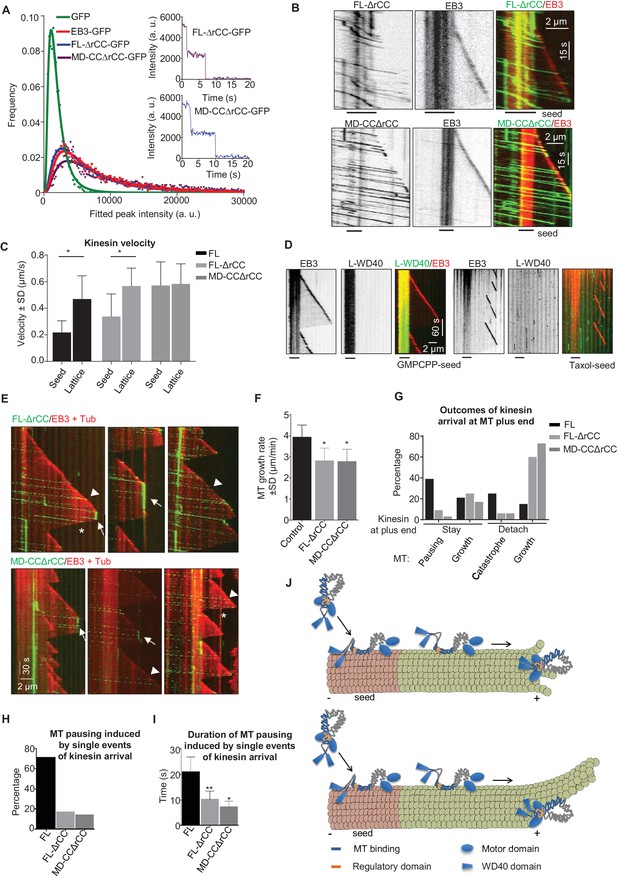
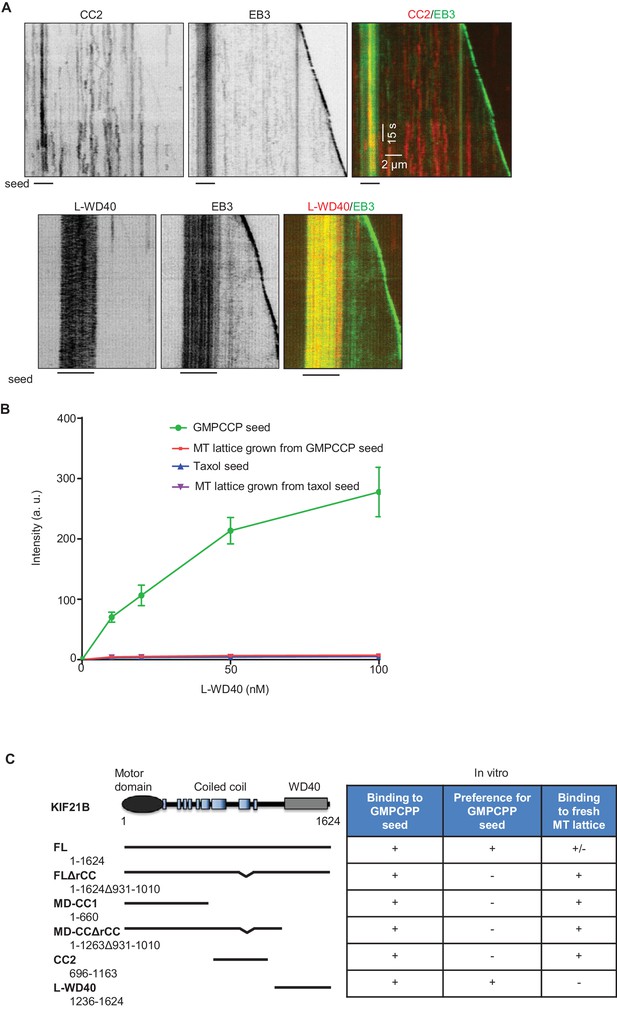
(A) Histograms of fluorescence intensities at the initial moment of observation of single molecules of the indicated proteins immobilized on coverslips (symbols) and the corresponding fits with lognormal distributions (lines). n = 3907, 5002, 6725 and 6943 molecules; fluorophore density was 0.19, 0.24, 0.30 and 0.33 µm−2 for GFP, GFP-EB3, KIF21B-FL-ΔrCC-GFP and KIF21B-MD-CCΔrCC-GFP proteins. Insets show representative photobleaching traces of individual molecules (background subtracted). (B) Kymographs illustrating the behavior of the indicated deletion mutants of KIF21B at 3 nM concentration on dynamic MTs in the presence of 100 nM mCherry-EB3. GMPCPP-stabilized MT seeds were labeled with Rhodamine-tubulin (lines below kymographs). Kymographs were generated from the movies acquired using Photometrics Evolve 512 EMCCD (Roper Scientific) camera (stream acquisition with an exposure time of 500 ms). (C) Quantification of the velocity of KIF21B-FL-GFP and the deletion mutants on seeds and freshly polymerized MT lattices, shown in Figures 3A and 7B. Seed: n = 295 for KIF21B-FL-GFP, n = 195 for KIF21B-FL-ΔrCC-GFP, n = 434 for KIF21B-MD-CCΔrCC-GFP; lattice: n = 131 for KIF21B-FL, n = 133 for KIF21B-FL-ΔrCC-GFP, n = 434 for KIF21B-MD-CCΔrCC-GFP. Data are from two or three independent experiments. Values significantly different from each other are indicated by asterisks, p<0.0001, Mann-Whitney U test. (D) Kymographs illustrating the interaction of purified GFP-L-WD40 (100 nM) with dynamic MTs grown from Rhodamine-tubulin labeled GMPCPP- or taxol-stabilized seeds (as indicated) in the presence of 20 nM mCherry-EB3. Kymographs were generated from the movies acquired in stream acquisition mode with an exposure time of 500 ms using Photometrics Evolve 512 EMCCD camera (Roper Scientific). (E) Kymographs illustrating the behavior of KIF21B deletion mutants on dynamic MTs in the presence of 20 nM mCherry-EB3 and 3% Rhodamine-tubulin. Pauses and KIF21B detachment from a depolymerizing MT end are indicated by arrows and asterisks, respectively. Arrowheads indicate kinesin detachment from the growing MT tip. Kymographs were generated from the movies acquired using CoolSNAP HQ2 CCD camera (Roper Scientific) with a 1.2-s interval between frames and an exposure time of 100 ms. (F) Quantification of MT growth rate in vitro in the presence of 15 µM tubulin with 20 nM mCherry-EB3 alone (n = 71) or together with 3 nM KIF21B-FL-ΔrCC-GFP (n = 79) or KIF21B-MD-CCΔrCC-GFP (n = 79). MTs were grown in the presence of 3% Rhodamine-tubulin. two independent experiments. (G) Quantification of different events observed after KIF21B-FL or its mutants reach a growing MT plus end. Data shown in Figure 3E are included here for comparison. n = 501 for KIF21B-FL-ΔrCC-GFP, n = 647 for KIF21B-MD-CCΔrCC-GFP. Data are from at least two independent experiments. (H) Percentage of pausing events induced by a single event of kinesin arrival from all detected pauses. Total number of pausing events: n = 51 for KIF21B-FL-GFP, n = 46 for KIF21B-FL-ΔrCC-GFP, n = 22 for KIF21B-MD-CCΔrCC-GFP. Data are from at least two independent experiments. (I) Quantification of the duration of MT pausing induced by a single kinesin arrival event at the growing MT plus end. n = 36 for KIF21B-FL-GFP, n = 8 for KIF21B-FL-ΔrCC-GFP, n = 3 for KIF21B-MD-CCΔrCC-GFP. Data are from at least two independent experiments.**p<0.0001, *p<0.0004 Mann-Whitney U test. (J) Model for the regulation of KIF21B motility and pause induction by the tail domain. In solution, KIF21B motor domains are inhibited by the regulatory region, while the WD40 domains are available for the interaction with MTs; WD40 domains show preference for the GMPCPP-stabilized seeds (red). After binding to seeds, KIF21B becomes activated and can walk to the plus end; it is likely that both the WD40 and the CC2 region contribute to MT binding. The kinesin can transfer from the seed to the freshly polymerized MT lattice; the interaction of the CC2 but not of the WD40 with the lattice promotes motor processivity. At the tip, the conversion to the autoinhibited conformation and the WD40 domain can prevent KIF21B from stepping off the MT plus end. This allows the motor to prevent both elongation and shortening of a small number of protofilaments with which it interacts. The remaining protofilaments might undergo short excursions of growth and shrinkage (upper panel); alternatively, they might elongate for some time and such an incomplete MT will be prone to bending and catastrophe (lower panel).
-
Figure 7—source data 1
An excel sheet with numerical data on the quantification of KIF21B mutants dimer analysis, photobleaching step analysis, velocities on seeds and MT lattices, MT growth rate in vitro and outcomes of the arrival of KIF21B mutants at MT plus ends, represented as plots in Figure 7A,C,F–I.
- https://doi.org/10.7554/eLife.24746.036
Characterization of KIF21B tail fragments in vitro
(A) In vitro reconstitution of MT dynamics in the presence of 100 nM GFP-EB3 and the extracts of HEK293T cells expressing mCherry-CC2 or mCherry-L-WD40. GMPCPP-stabilized MT seeds were labeled with HiLyte Fluor 488-tubulin (lines below kymographs). (B) Quantification of the intensity of purified GFP-L-WD40 on GMPCPP- and taxol-stabilized seeds or dynamic MTs grown from GMPCCP or taxol-stabilized seeds. (C) Overview of the interactions of GFP-tagged KIF21B and its deletion mutants with MTs in vitro.
-
Figure 7—figure supplements 1—source data 1
An excel sheet with numerical data on the quantification of the intensity of KIF21B-L-WD40 on seeds and dynamic MTs represented as plot in Figure 7—figure supplement 1B.
- https://doi.org/10.7554/eLife.24746.038
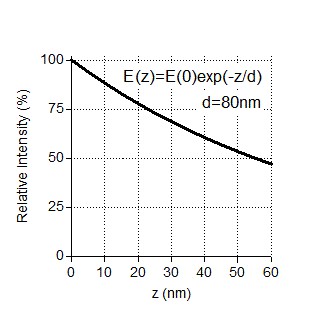
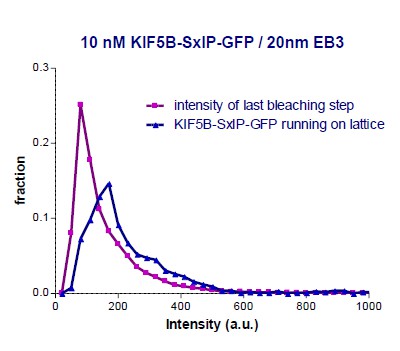

Example image of KIF21B-FL-GFP density used for quantification.
It can be seen that the density of single molecules is in the optimal range
Videos
KIF21B induces pausing of a depolymerizing MT.
The movie shows the arrival of KIF21B-FL-GFP at the end of a depolymerizing MT and a subsequent pausing event. The arrival of additional KIF21B-FL-GFP molecules results in a long pause. The experiment was performed in the presence of 15 µM tubulin, Rhodamine-tubulin (0.5 µM), mCherry-EB3 (20 nM) and KIF21B-FL-GFP (0.5 nM). The movie consists of 200 frames acquired with a 1.2-s interval between frames and an exposure time of 100 ms. Scale bar, 2 μm.
KIF21B perturbs MT growth and induces MT bending.
The combined movie shows the two different events illustrated in Figure 4—figure supplement 1A and Figure 4B. The movie shows bending of an MT growing beyond the point where KIF21B-FL-GFP was stalled (top panel, upper MT) and repeated short excursions from the point of KIF21B-FL-GFP stalling (bottom panel). The experiment was performed in the presence of 15 µM tubulin, Rhodamine-tubulin (0.5 µM), mCherry-EB3 (20 nM) and KIF21B-FL-GFP (0.5 nM). The movie consists of 128 frames acquired with a 1.2-s interval between frames and an exposure time of 100 ms. Scale bar, 2 μm.
Additional files
-
Supplementary file 1
Analysis of purified KIF21B and its deletion mutants used in this study by mass spectrometry.
Samples of purified KIF21B proteins were loaded on SDS-PAGE, isolated from the gel after in-gel digestion and subsequently analyzed by mass spectrometry to test for purity. All identified proteins are included in Supplementary file 1in alphabetical order. Indicated are the molecular weight and the number of unique peptides found for identified proteins in the different KIF21B samples. In total, 121 proteins were identified for KIF21B-FL-GFP, 183 for KIF21B-FL-ΔrCC-GFP, 107 for KIF21B-MD-CCΔrCC-GFP, 92 for GFP-L-WD40 and 63 for KIF21B-MD-CC1-GFP.
- https://doi.org/10.7554/eLife.24746.039
-
Supplementary file 2
Lognormal (best fit) values for the fluorescence intensity measurements.
- https://doi.org/10.7554/eLife.24746.040





