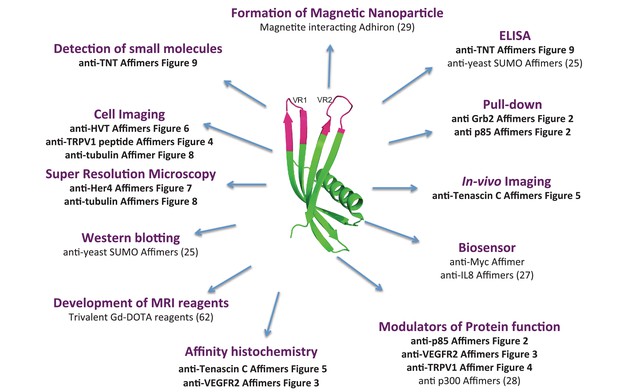
Affimer proteins are versatile and renewable affinity reagents
Figures
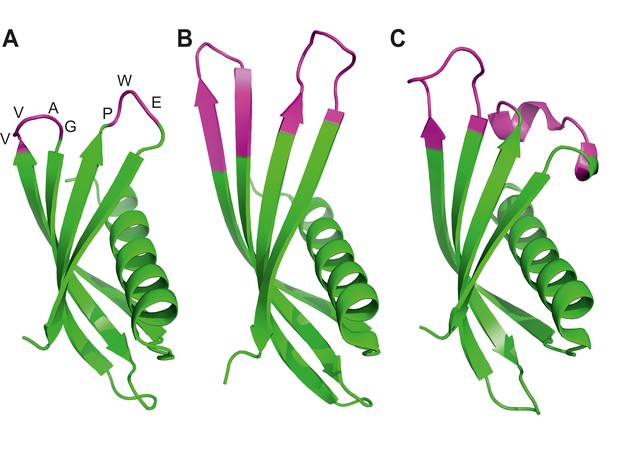
Ribbon diagrams of three crystal structures for Affimer (Adhiron) reagents.
(A) X-ray crystal structure of Affimer scaffold (PDB ID no. 4N6T) at 1.75 A resolution. The amino acids from the loops connecting the four anti-parallel beta sheets are highlighted in pink. (B) Crystal structure of an Affimer against p300 (PDB ID no. 5A0O) (C) Crystal structure of an Affimer isolated against human SUMO proteins (PDB ID no. 5ELJ). The variable regions in B and C are shown in pink.
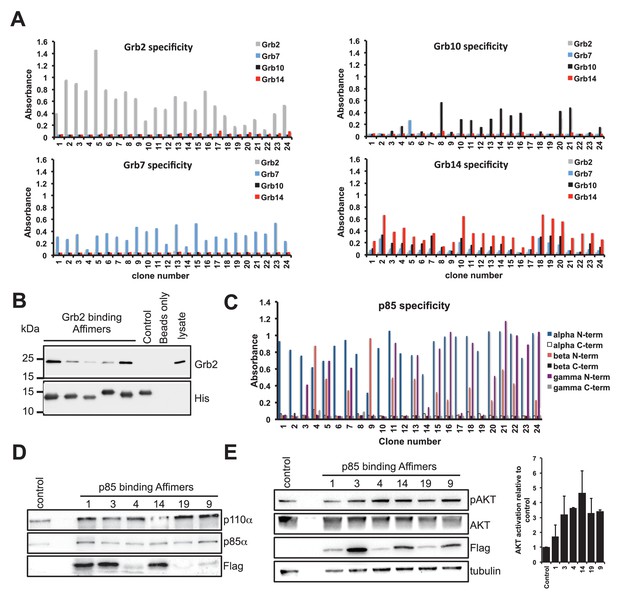
Isolation and characterisation of SH2 domain binding Affimers.
(A) Phage ELISA from 24 monoclonal Affimer reagents isolated against the respective Grb family member SH2 domains. Specificity was tested through extent of binding to the other SH2 family members. (B) Western blot showing Affimer-mediated affinity-precipitation of endogenously expressed Grb2 protein from U2OS cell lysates using five Grb2 Affimers bound to colbalt magnetic beads (n = 2). A yeast SUMO binding Affimer was used as a negative control. (C) Phage ELISA from 24 monoclonal Affimer reagents isolated against p85 alpha N-terminal domain family member SH2 domain. Specificity was tested through extent of binding to the other p85 SH2 family members. (D) Western blot of immunoprecipitation using a p110 antibody on cell lysates from cells expressing p85 SH2 domain binding Affimers (n = 3). (E) Western blot and quantification by densitometry of AKT phosphorylation in the presence of expressed p85 SH2 domain binding Affimers (n = 2).
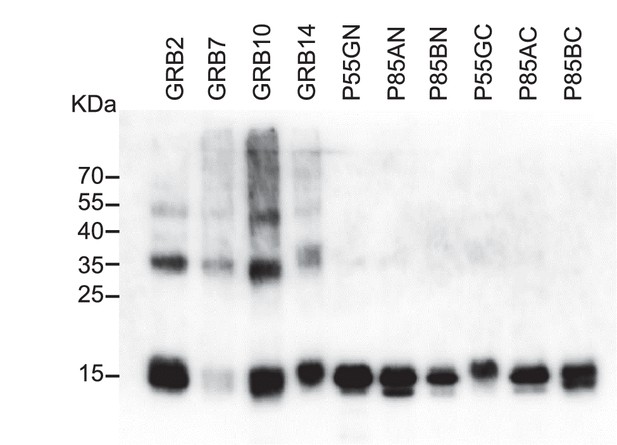
Western blot results of Avi-Tag SH2 domain proteins using an streptavidin-HRP conjugate to detect the presence of biotin.
https://doi.org/10.7554/eLife.24903.005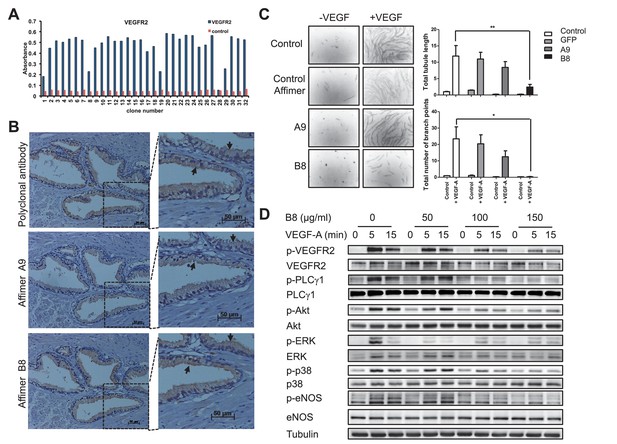
Characterisation of VEGFR2 binding Affimers.
(A) Phage ELISA for 32 monoclonal Affimer reagents isolated against VEGFR2. The negative control contained just streptavidin. (B) Immuno- and affinity-histochemistry of a polyclonal anti-VEGFR2 antibody and of representative Affimers B8 and A9. Staining is shown as a light brown color, haemotoxylin counter staining (blue). Arrows show similar staining patterns. (C) Tubulogenesis assay in the presence and absence of vascular endothelial growth factor A and the two Affimers with quantification of tubule length and branch point number shown to the right. The control is in the absence of any Affimer and the control Affimer is a binder against yeast SUMO (n = 3). Statistical analysis was performed using a two-way ANOVA followed by the Bonferroni multiple comparison test using GraphPad Prism software (La Jolla, USA). p values less than 0.05 (*), 0.01 (**) are indicated on the graphs. Error bars in graphs denote ± standard error of mean. (D) Western blot results showing changes in downstream signalling in HUVECs treated for 0, 5 and 15 min in the presence of vascular endothelial growth factor A and increasing concentrations of the VEGFR2 binding Affimer B8 (n = 3).
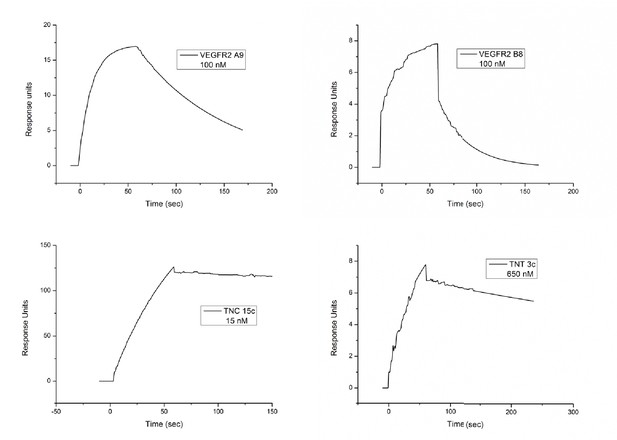
SPR plots for the anti-VEGFR2, TNC and TNT binding Affimers.
https://doi.org/10.7554/eLife.24903.007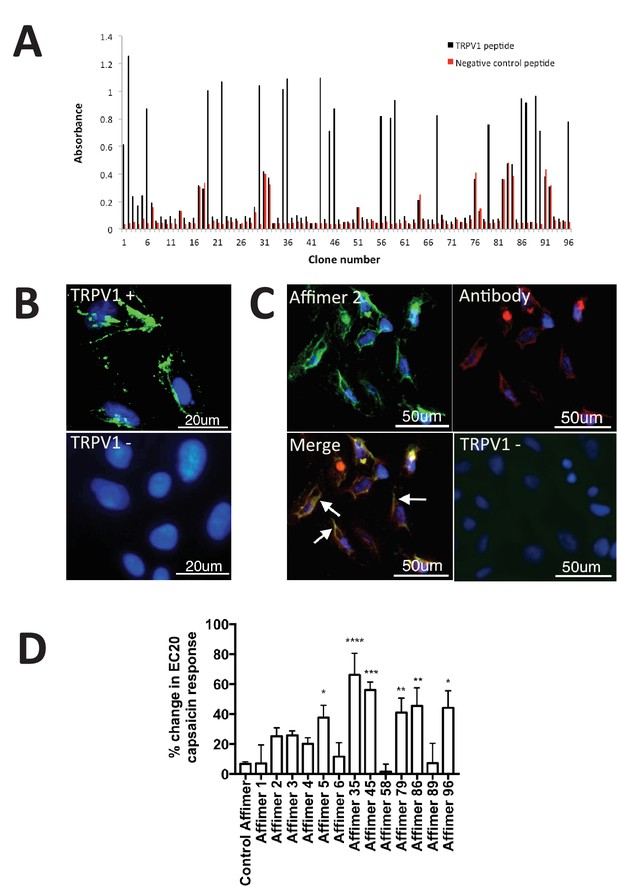
Characterisation of TRPV1 binding Affimers.
(A) Phage ELISA for 96 monoclonal Affimer reagents isolated against TRPV1 peptide. The negative control contained a different hydrophobic peptide sequence. (B) Affinity-cytochemistry on U2-OS cells transiently transfected with TRPV1 (TRPV1+) or control (TRPV1-) using Affimer 2. Binding was detected using an anti-HIS antibody fluorescently labeled with FITC. Binding of the Affimer is shown as a green and DAPI (a DNA stain) shown as blue (n = 3), (C) Co-localisation of Affimer staining with an anti-TRPV1 antibody. Antibody staining is shown in red. (D) A Flexstation was used to measure uptake of Fluo-4 AM, a calcium binding fluorescent small molecule, to measure calcium levels in capsaicin stimulated cells in the presence of Affimer control and TPRV1-binding Affimers (n = 3).
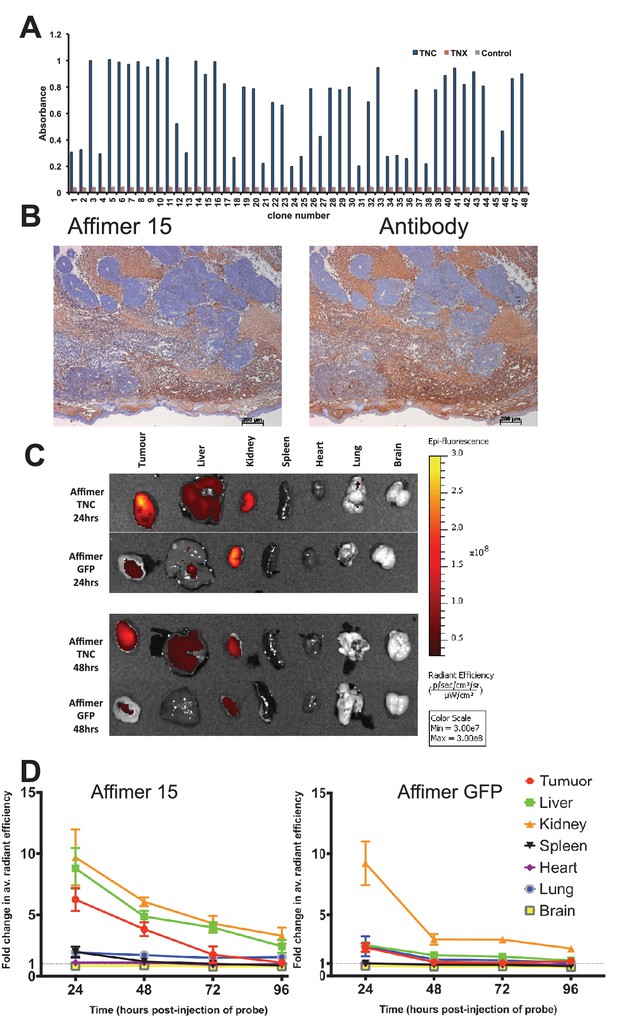
Characterisation of tenascin C (TNC) binding Affimer by affinity-histochemistry and ex vivo imaging of xenografts.
(A) Phage ELISA for 48 monoclonal Affimers against TNC. The two controls are tenascin X (TNX) and streptavidin. (B) Immunohistochemistry of serial sections of a mouse xenograft (SW620 cell line), showing staining for TNC. Antibody/Affimer staining is shown as a light brown color with haemotoxylin counter staining (blue). (C) and (D) Mice were injected via their tail vein with rhodamine labelled TNC binding Affimer or a control GFP binding Affimer. After 24, 48, 72 and 96 hr the xenograft and organs were removed and visualized. (C) Organ images at 24 hr. (D) Quantification of rhodamine fluorescence (radiant efficiency in p/s/cm2/sr/μW/cm2) ex vivo (n = 3). Mean background fluorescence intensity was normalized to sham injected control tumors and organs.
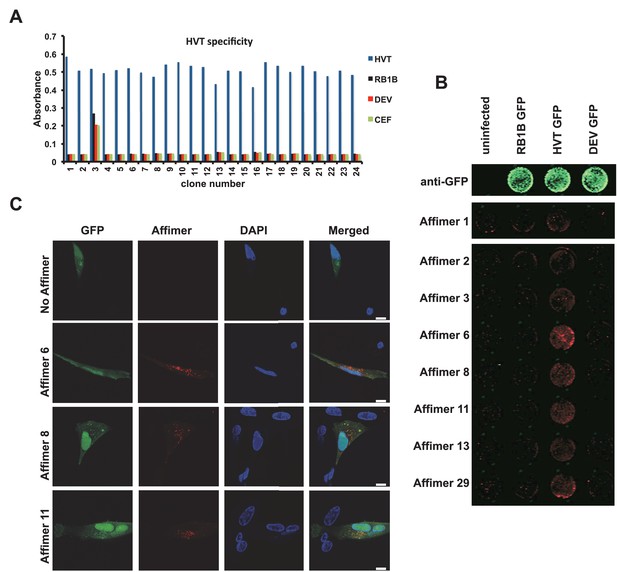
Affimer detection of HVT UL49 in infected cells by in-cell Western and affinity-fluorescence.
(A) Phage ELISA for 24 monoclonal Affimers against HVT screened against HVT, RB1B, DEV and CEF lysates to confirm specificity for HVT. (B) Infection of cells was confirmed using a goat anti-GFP antibody and donkey anti-goat 680 (green) antibody to detect GFP which is constitutively expressed by the BAC derived viruses. Infected CEFs were screened with candidate HVT UL49 Affimers at 1.5 µg/ml with subsequent detection (red) by Streptavidin 800 conjugate (Licor). (n = 3) (D) HVT infected CEFs were screened with HVT UL49 Affimers and visualised with Streptavidin-568 (red). Nuclei were stained with DAPI (blue). Streptavidin only control (no Affimer) shows no observable labelling of infected CEFs. Bar = 10 µm.
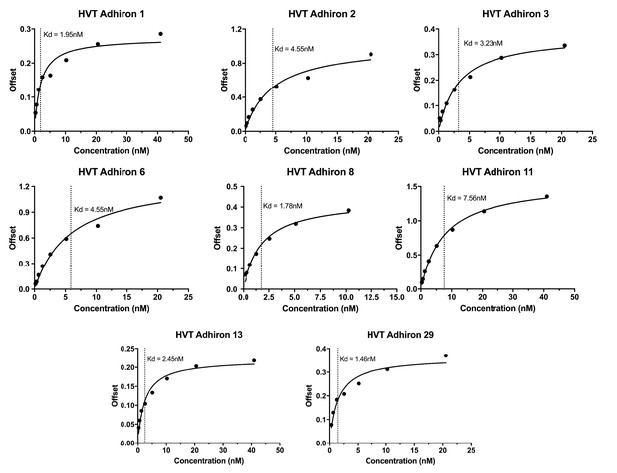
Bilayer Interferometry plots for the HVT binding Affimers.
https://doi.org/10.7554/eLife.24903.011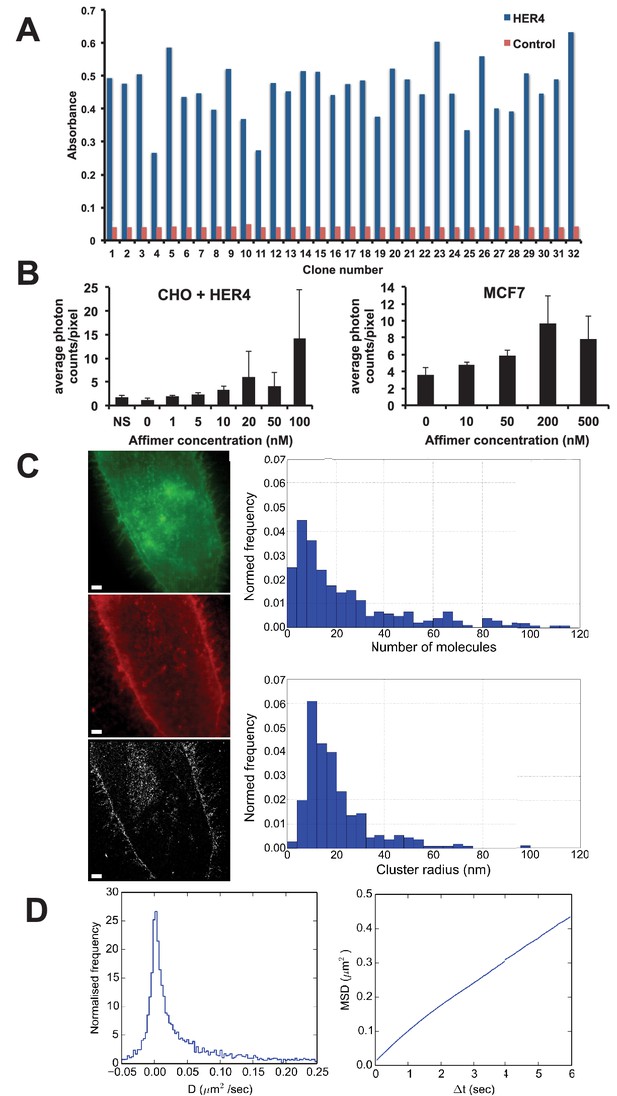
Use of HER4 binding Affimers in super-resolution imaging and single molecule tracking.
(A) Phage ELISA for HER4 binding Affimers. (B) Average photon counts/pixel for HER4-binding Affimer labelled with CF640R and bound to CHO cells transfected with HER4 and to MCF7 cells expressing endogenous levels of HER4. (C) Wide field image of CHO cells transfected with HER4-CYT-eGFP showing localisation of HER4 via GFP fluorescence (top) and labelled with HER Affimer–Alexa647 (middle). The corresponding dSTORM image of HER4 Affimer conjugated to Alexa647 (bottom) with a 25 nm localisation precision. Scale bar = 2 μm. Right plots to show the number of molecules and cluster size of clusters identified by dSTORM. (D) Diffusion coefficients (left panel), and MSD curve (right panel) of HER4 Affimers labelled with CF640R and tracked on MCF7 cells expressing endogenous HER4.
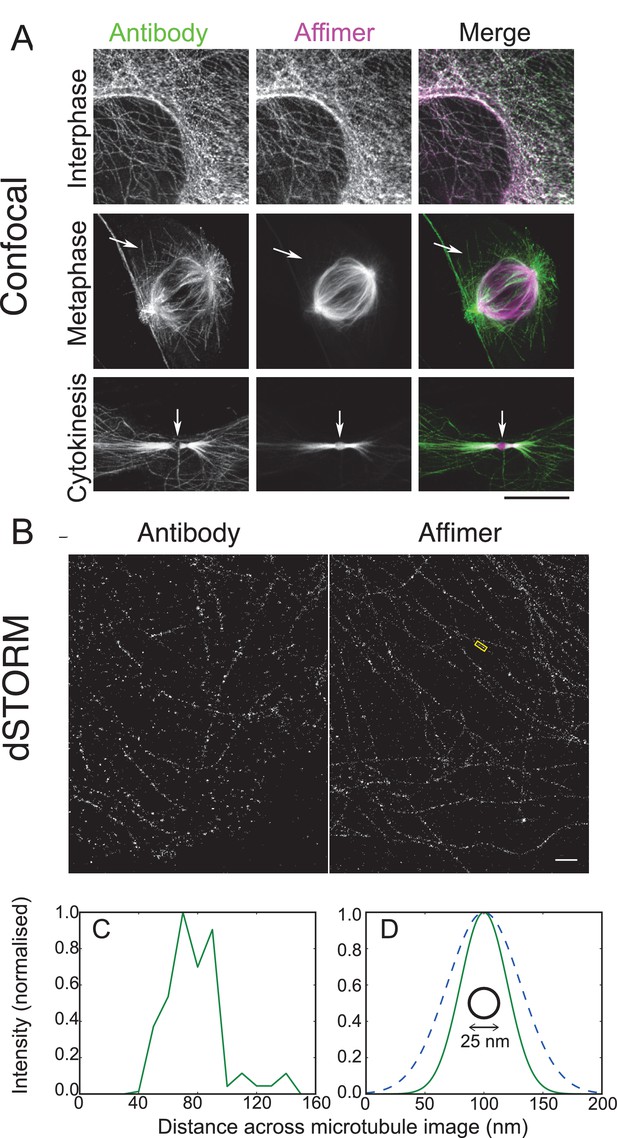
Use of tubulin binding Affimer in super-resolution microscopy.
(A) Confocal images of microtubules in HeLa cells, stained with a rat α-tubulin antibody (YL1/2) which recognises tyrosinated tubulin, and an Affimer for polymerised tubulin, conjugated to Alexa Fluor 647. Images of an interphase and metaphase cell, together with an image of the cytokinetic furrow are shown. Arrows in the metaphase cell point to astral microtubules that are predominantly labelled with the antibody. Arrows in the cyokinetic furrow indicate the central region (Fleming body). Scale bar is 10 μm. (B) 3D dSTORM images of microtubules in a HeLa cell, labelled with Alexa Fluor 647 conjugated to a primary antibody to rat α-tubulin (left) and an Affimer for polymerised tubulin (right). These images are from separate cells. Localisations were aggregated into 10 nm bins and projected onto a single plane, with Gaussian smoothing. Scale bar 1 µm. (C) Intensity profile across the microtubule image labelled in (B) (yellow box), averaged along 510 nm of its length. The central decrease in intensity reflects the hollow structure of the microtubule. (D) Comparison of the average microtubule image intensity profile with antibody staining (dashed, mean of 6 microtubule sections), Affimer staining (solid, mean of 8 microtubule sections) and actual microtubule size (black circle). The FWHM of each average profile (as in (C)) was found for a Gaussian fit and a Gaussian distribution is plotted here using the mean FWHM for each staining method.
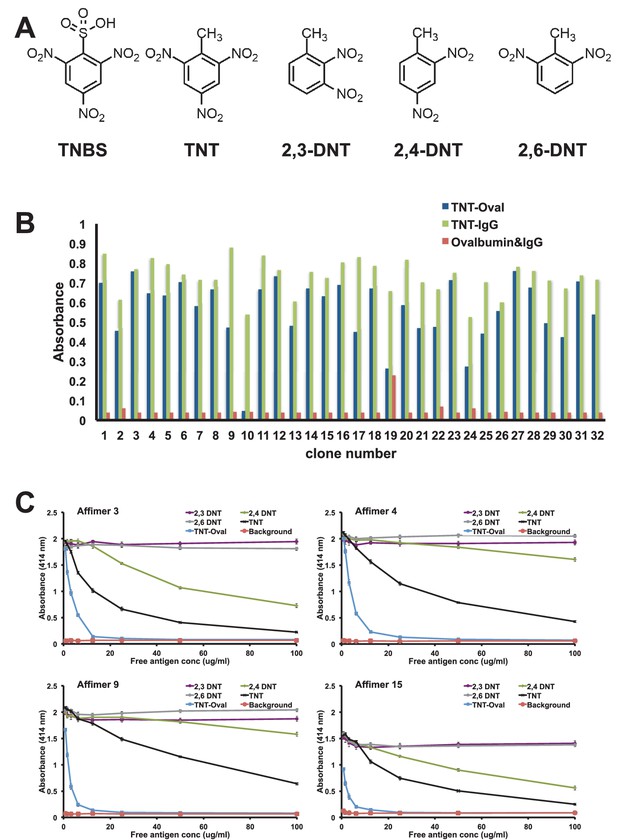
Affimer selection and specificity against TNT and DNT’s.
(A) Chemical structures of TNBS, TNT, 2,3-DNT, 2,4-DNT and 2,6-DNT. (B) Phage ELISA results from 32 monoclonal Affimer reagents isolated against TNBS bound to ovalbumin. Binding specificity was also tested against TNBS bound to IgG and to unconjugated ovalbumin and IgG. (C) Competition ELISA of four TNT-Affimers to check specificity against a range of molecules across a concentration profile. Error bars = standard deviation from technical repeats of a representative ELISA.