Systematic bacterialization of yeast genes identifies a near-universally swappable pathway
Figures
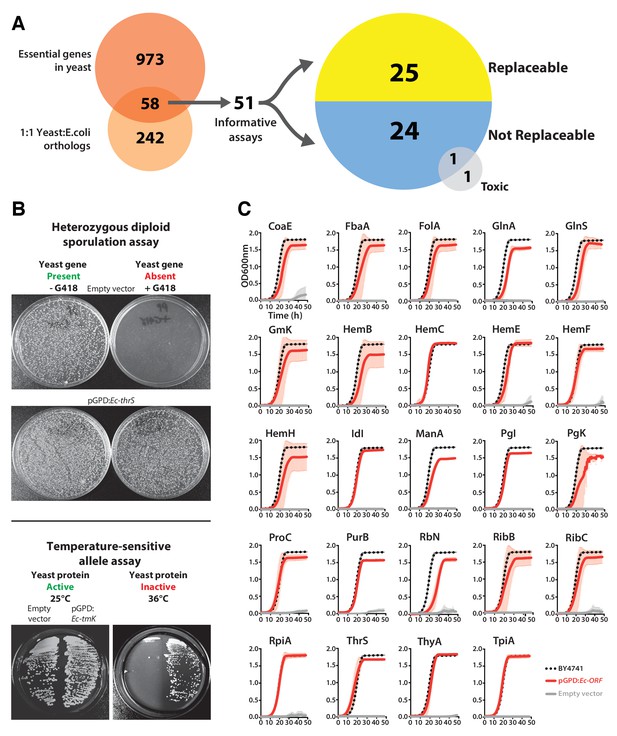
Many E. coli genes efficiently complement lethal growth defects in their yeast counterparts.
(A) Yeast and E. coli share hundreds of genes, 58 of which are essential in yeast and have clear 1:1 orthologs in either species. E. coli genes were cloned into a yeast expression vector under the control of a GPD promoter. 51 of these 58 E. coli genes provided informative assays for replaceability in yeast. Initial results from these complementation assays revealed that 25 of 51 (~49%) E. coli genes could functionally replace their orthologous yeast counterparts. (B) Complementation assays were performed in two different yeast strain backgrounds, as shown for representative assays. In the case of a yeast strain with a temperature-sensitive allele of the yeast gene Sc-cdc8, cells carrying the empty vector control grow at the permissive-temperature (25°C, yeast protein active) but not the restrictive-temperature (36°C, yeast protein inactive), unlike cells expressing the E. coli ortholog (Ec-tmK), indicating that the E. coli gene can functionally replace the yeast gene. In the case of yeast heterozygous diploid (Sc-ths1Δ/Sc-THS1) deletion strain, cells are sporulated and haploid progeny grown on selective medium (-Ura -Arg -His -Leu + Can) in the absence (yeast gene present) or presence of G418 (200 μg/ml) (yeast gene absent). Cells expressing the E. coli ortholog (Ec-thrS) grow on G418-containing medium, unlike cells carrying the empty vector control, indicating successful complementation. (C) Haploid yeast gene deletion strains carrying plasmids expressing functionally replacing E. coli genes (red solid-lines) generally exhibit comparable growth rates to the wild type parental yeast strain BY4741 (black dotted-lines). The empty vector control (grey solid-line) showed no such growth rescue in the presence of G418. Mean and standard deviation plotted with N = 3.
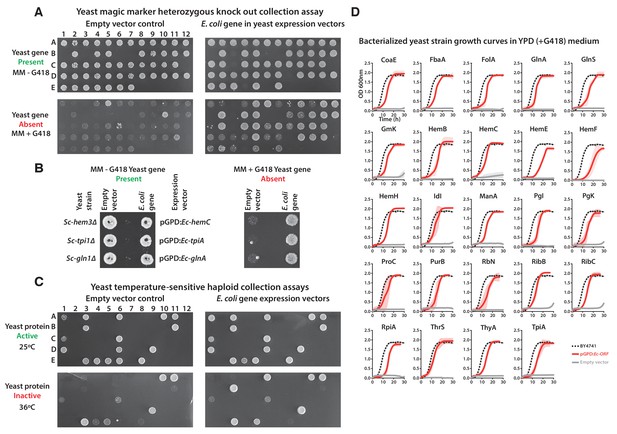
Complementation assays performed in a 96-well format in two different yeast strain backgrounds (Supplementary file 1).
(A and B) Magic marker heterozygous diploid deletion yeast strains expressing E. coli genes were sporulated and the sporulation mix was spotted on magic marker agar medium (-Ura -Arg -His -Leu + Can) with (yeast gene absent) or without (yeast gene present) G418 (200 μg/ml). (C) Temperature-sensitive haploid yeast strains expressing E. coli genes grown at permissive temperature (25°C) (yeast protein active) and at restrictive temperature (36°C) (yeast protein inactive) on -Ura agar medium with G418 (200 μg/ml). Empty vector containing yeast cells were used as negative control for the experiment. (D) Haploid yeast gene deletion strains carrying plasmids expressing functionally replacing E. coli genes (red solid-lines) generally exhibit comparable growth rates to the wild type parental yeast strain BY4741 (black dotted-lines) as grown in YPD liquid medium in the presence of G418 (300 μg/ml). Mean and standard deviation plotted with N = 3.
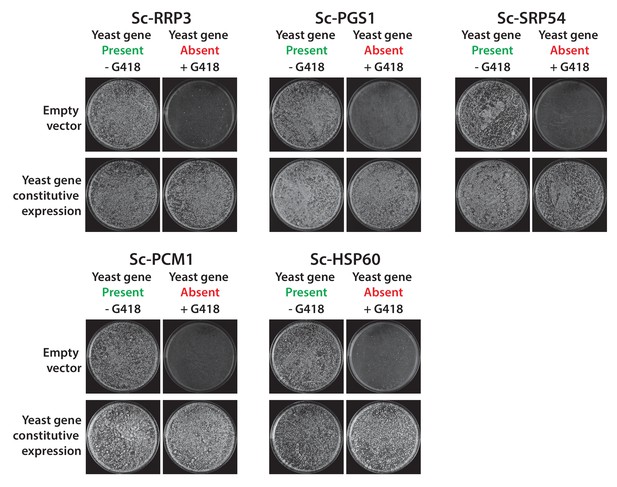
Constitutive plasmid expression of yeast genes efficiently replaced the corresponding genomic copies for 6 non-replaceable alleles.
Bacterial orthologs of the yeast genes, Sc-RRP3, Sc-PGS1, Sc-SRP54, Sc-PCM1 and Sc-HSP60 did not show functional replacement when expressed from a constitutive GPD promoter. We expressed the corresponding yeast genes in a similar fashion under the control of the constitutive GPD promoter. All the tested yeast genes functionally replaced the corresponding yeast gene deletions. Empty vector containing yeast cells were used as negative control for the experiment.
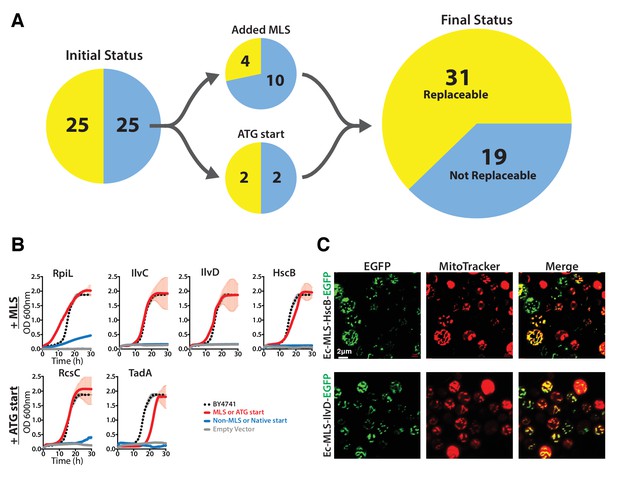
The addition of a mitochondrial localization signal (MLS) and mutation of start codons from GTG to ATG allows some E. coli genes to swap for their respective yeast orthologs.
(A) 14 of the 25 non-replaceable E. coli genes were predicted to function in mitochondria in yeast. 4 of 14 were replaceable after adding the MLS at the N-termini of the E. coli genes. Site-specific mutagenesis of E. coli gene start codon from GTG to ATG allowed two to functionally complement the corresponding yeast genes bringing the total number E. coli genes that functionally replace yeast genes to 31 of 51 (~61%). (B) Haploid yeast gene deletion strains carrying mitochondrially localized E. coli genes rescued the growth defect of the yeast gene (red solid-line) comparable to the wild type yeast (black dashed-line). The empty vector control (grey solid-line) and the yeast cells expressing of E. coli gene without MLS (blue-solid line) showed no such growth rescue in the presence of G418. Mean and standard deviation plotted with N = 3. (C) EGFP-tagged E. coli genes that functionally replaced the yeast gene function were imaged after MitoTracker red staining. EGFP-tagged Ec-MLS-HscB and Ec-MLS-IlvD (green) show colocalization with MitoTracker red stained mitochondria (red).
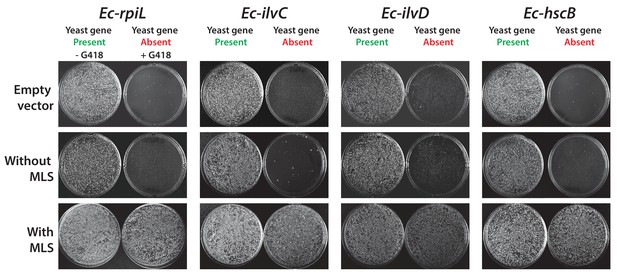
Some E. coli genes require a yeast mitochondrial localization signal to efficiently replace.
The magic marker heterozygous diploid deletion yeast strains carrying empty vector or E. coli gene with or without MLS were sporulated and the sporulation mix was plated on magic marker agar medium (-Ura -Arg -His -Leu + Can) with or without G418 (200 μg/ml). E. coli genes Ec-rpiL, Ec-ilvC, Ec-ilvD and Ec-hscB without an appropriate mitochondrial localization signal cannot complement the corresponding yeast gene deletions Sc-mnp1, Sc-ilv5, Sc-ilv3 and Sc-jac1. However, expression of E. coli genes with yeast MLS efficiently rescued the growth defect of the corresponding yeast gene deletions.
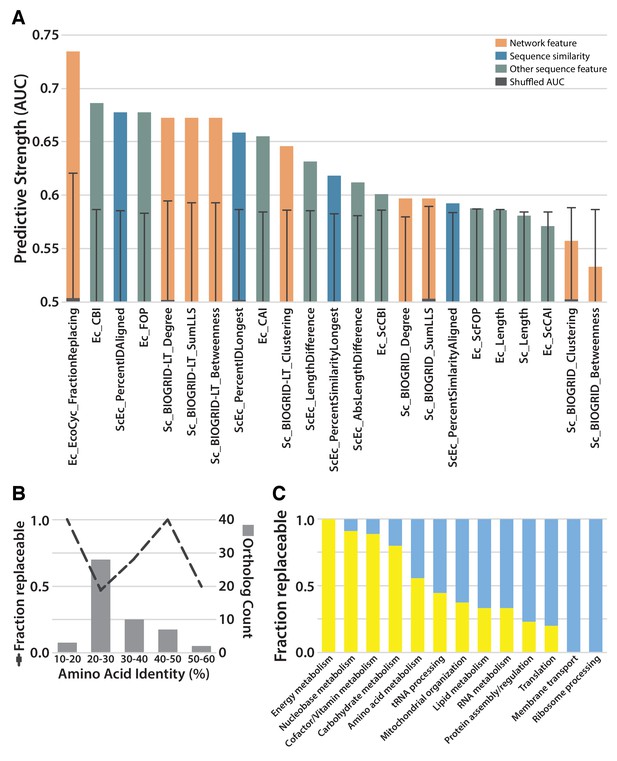
Replaceability of E. coli genes is a modular phenomenon.
(A) Several quantitative properties of the tested genes were assessed for their ability to predict replaceability, measured as the area under a receiver operating characteristic curve (AUC). Having a high fraction of interaction partners that replace was the most predictive property tested, suggesting that the ability to replace is a modular phenomenon whereby genes functioning together are similarly able to replace. A Random Forest classifier constructed with all attributes boosted the maximum AUC to 0.79. (B) As shown in (A), sequence similarity was not the most predictive feature. The fraction of replaceable genes in given ranges of similarity was variable, with the vast majority of orthologs being 20–30% identical, a range in which roughly half of proteins replaced. (C) Mapping of replaceability status onto yeast GO slim annotations revealed that GO categories have varying rates of replaceability, with core metabolic processes (e.g. energy metabolism, nucleobase metabolism) being largely replaceable while more specialized processes (e.g. protein assembly, membrane transport) were less so.
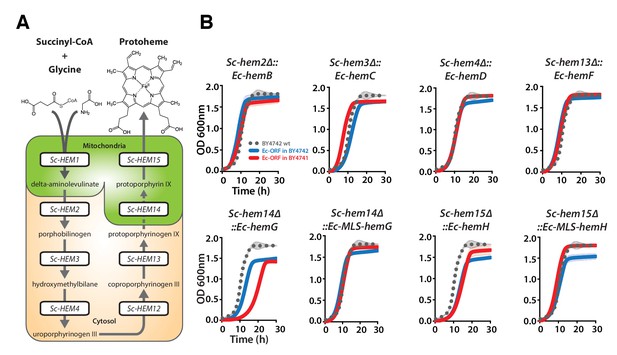
Bacterialization of yeast heme biosynthesis pathway genes at their native loci.
(A) A schematic of the yeast heme pathway shows the beginning of the pathway in mitochondria using succinyl-CoA and glycine as precursors. The subsequent enzymatic reactions are cytosolic up until the penultimate and ultimate reactions which are mitochondrial. (B) Growth kinetics of CRISPR-Cas9 engineered yeast heme pathway genes replaced with the corresponding bacterial genes at their native yeast loci show efficient replaceability in both BY4741 (red solid-line) and BY4742 (blue solid-line) yeast strains. The wild type BY4741 growth curve is shown as a comparison (black dotted-line). Mean and standard deviation plotted with N = 3.
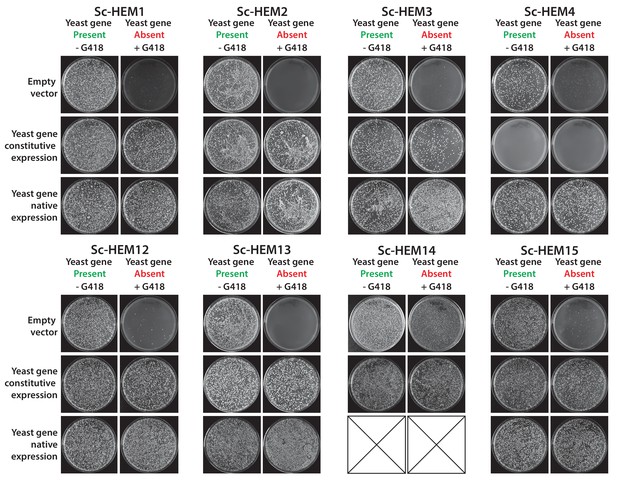
Constitutive or native plasmid-based expression of the yeast heme biosynthesis genes generally efficiently complemented growth defects in the corresponding yeast gene deletion strains.
Heterologous expression of yeast genes Sc-HEM1, Sc-HEM2, Sc-HEM3, Sc-HEM4, Sc-HEM12, Sc-HEM13, Sc-HEM14 and Sc-HEM15 under the control of constitutive GPD promoter or native promoter efficiently rescued the growth defect of the corresponding yeast gene deletions respectively except in the case of Sc-HEM4. Sc-HEM4, when expressed constitutively, resulted in toxicity in the presence of the yeast gene at the native locus and did not complement the function in the absence of the yeast gene. This toxicity was relieved when the yeast gene was expressed under the control of the native yeast promoter.
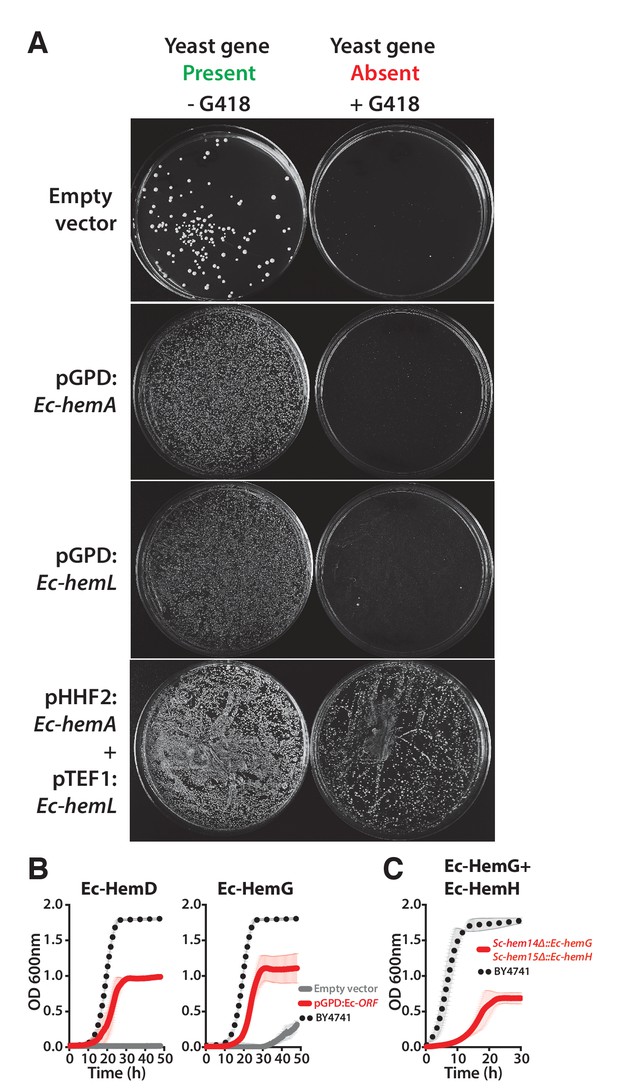
Ec-hemA and Ec-hemL carry out the initial reaction in E. coli heme biosynthesis and are both required to complement Sc-HEM1 deletion in yeast, and non-orthologous yeast genes are replaced by E. coli genes that carry out the identical reaction.
(A) Expression of heme pathway genes of E. coli, Ec-hemA or Ec-hemL, individually cannot complement the lethal growth defect of the deletion of Sc-HEM1 gene in yeast. Co-expression of Ec-HemA and Ec-HemL efficiently rescued the growth defect of Sc-hem1 gene deletion in yeast. (B) Growth curves of yeast strains with deletions of Sc-hem4 and Sc-hem14 genes (grey solid-line) show functional replaceability (red solid-line) by the non-orthologous E. coli genes Ec-hemD and Ec-hemG that carry out identical enzymatic reactions to the corresponding yeast genes. The wild type BY4741 growth curve is shown as a comparison (black dotted-line). The empty vector control (grey solid-line) showed no such growth rescue in the presence of G418. (C) Growth curve of engineered yeast strain Sc-hem14Δ::Ec-hemG; Sc-hem15Δ::Ec-hemH in YPD medium harboring E. coli genes at the native yeast loci. The strain displayed a growth defect (red solid-line) compared to the wild type BY4741 strain (black dotted-line). Mean and standard deviation plotted with N = 3.
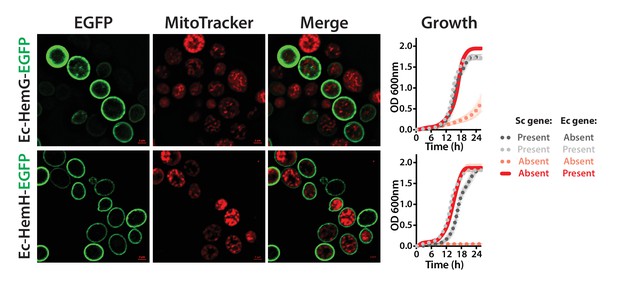
The penultimate and ultimate heme pathway enzymes in yeast are replaceable by their bacterial orthologs, in spite of mis-localizing to the plasma membrane.
EGFP-tagged Ec-HemG and Ec-HemH localize to the plasma membrane in yeast. The EGFP-tagged proteins do not localize to the mitochondria since no clear co-localization is observed with the Mitotracker red stain. EGFP-tagged Ec-HemG and Ec-HemH expression (red solid-line) efficiently rescue the growth defects of the respective yeast gene deletions (Sc-hem14 and Sc-hem15) (pink dotted-line) comparable to the wild type yeast (black dotted-line). Empty vector control is incapable of rescuing the growth defect of the deletion strains (grey dotted-line).
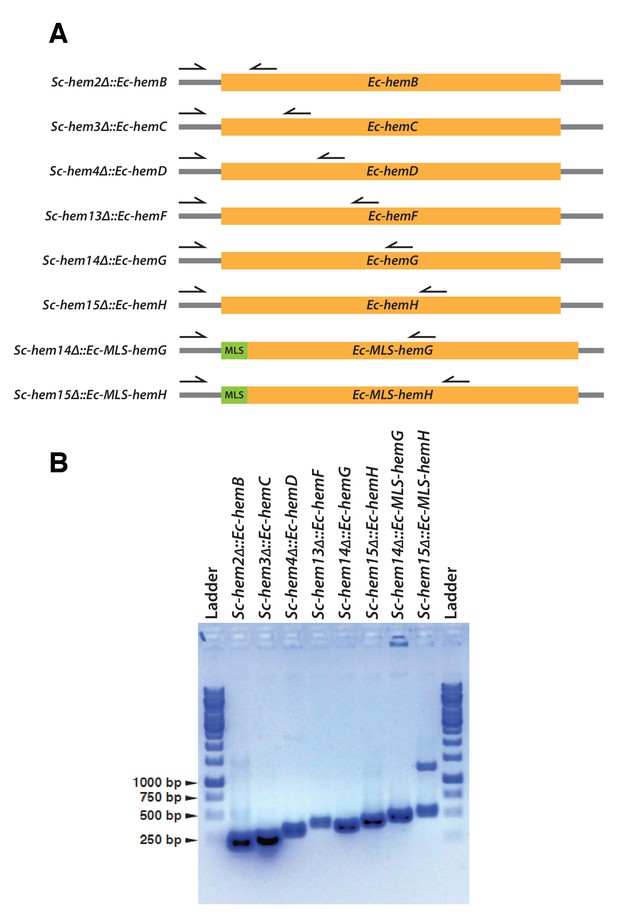
Confirmation of CRISPR-Cas9 mediated bacterialized yeast strains.
(A) Schematics of the yeast heme pathway gene loci carrying functionally replaceable E. coli genes while retaining their native promoters and terminators. The arrows indicate the primers used to confirm the replacement (refer to Supplementary file 3). (B) PCR amplification of expected size was obtained for each individual bacterialized yeast strains.
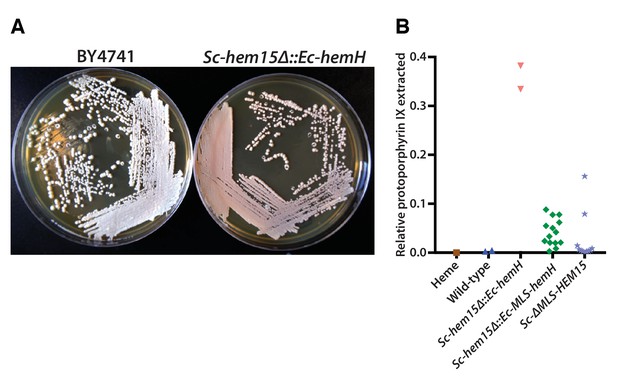
Mislocalization of the bacterialized ferrochelatase enzyme identifies a porphyria-like phenotype in yeast.
(A) Bacterialization of the ultimate yeast gene in the heme biosynthesis pathway results in a distinct pink colony phenotype on YPD agar medium. In contrast, wild type BY4741 strain colonies appear as creamy-white. (B) Acetate-extracted secreted products from the pink Sc-hem15Δ::Ec-hemH strains show strongly enhanced fluorescence at 635 nm (excitation 399 nm), comparable to a protoporphyrin IX standard and unlike a heme standard or extracts from the parental BY4741 strain. The introduction of an MLS to the bacterialized yeast strain (Sc-hem15Δ::Ec-MLS-hemH) significantly reduced protoporphyrin IX secretion, while deletion of the MLS from the native yeast locus in strain Sc-ΔMLS-HEM15 caused several strains to increase protoporphyrin IX secretion.
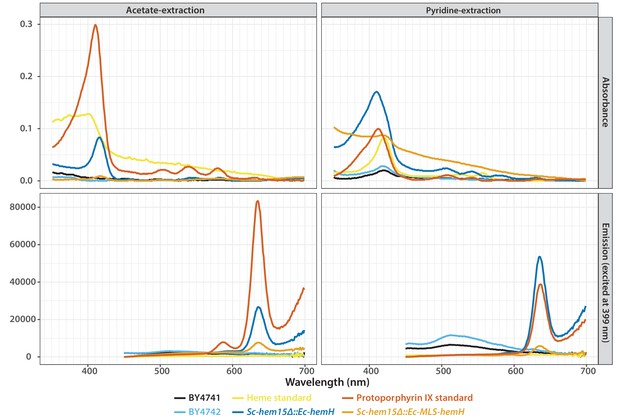
Absorbance (top) and emission (bottom) spectra of extracts obtained from acetate (left) and pyridine (right) extraction of the wild type or bacterialized yeast colonies grown on YPD medium.
Purified protoporphyrin IX (red solid-line) or heme (yellow solid-line) were used as standards. Extract from the bacterialized Sc-hem15Δ::Ec-hemH yeast strain (dark blue-line) matched with that of the protoporphyrin IX standard. Bacterialized ScHEM15Δ::Ec-MLS-hemH yeast strain (orange solid-line) showed significantly reduced peak for protoporphyrin IX. Extracts from wild type BY4741 (black-line) and BY4742 (light blue solid-line) were used as controls.
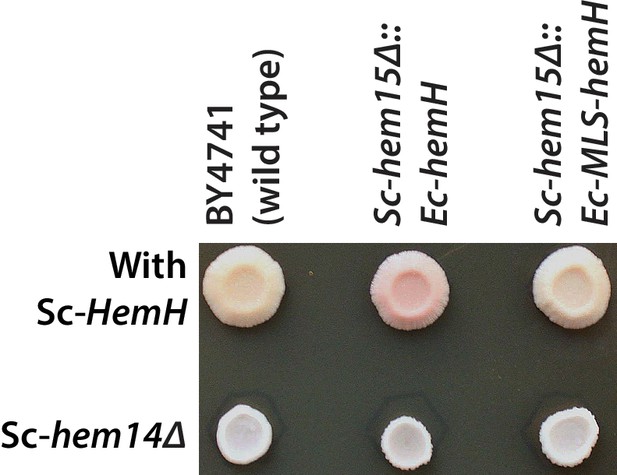
Deletion of protoporphyrinogen oxidase, Sc-HEM14, in the Sc-hem15Δ::Ec-hemH strain suppressed the porphyria-like pink phenotype.
Top row from left show growth spots of the BY4741 wild type, Sc-hem15Δ::Ec-hemH and Sc-hem15Δ::Ec-MLS-hemH yeast strains. Bottom row from left show corresponding strains harboring Sc-hem14 deletion.
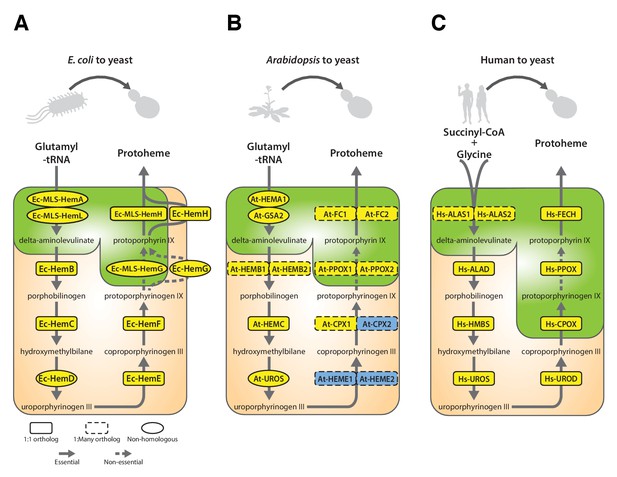
Yeast heme biosynthesis pathway enzymes can be successfully replaced by orthologs or analogs from bacteria, plants, and humans, in spite of alterations to subcellular localization.
Enzymatic steps of extant bacterial and eukaryotic heme biosynthesis pathways are identical save for the starting metabolites and conversion to delta-aminolevulinate; bacteria also exhibit non-orthologous gene displacement of several enzymes. Heme biosynthesis occurs in the bacterial cytoplasm and inner membrane, the human and yeast in mitochondria and cytoplasm, and the plant in chloroplast and cytoplasm. In spite of these localization changes over evolution, most of the defects in growth rate and viability conferred by heme pathway mutations in yeast can be complemented by introduction of the corresponding (A) bacterial genes, (B) plant genes (except for At-HemE), and (C) human genes. Yellow indicates a replaceable gene, blue non-replaceable.
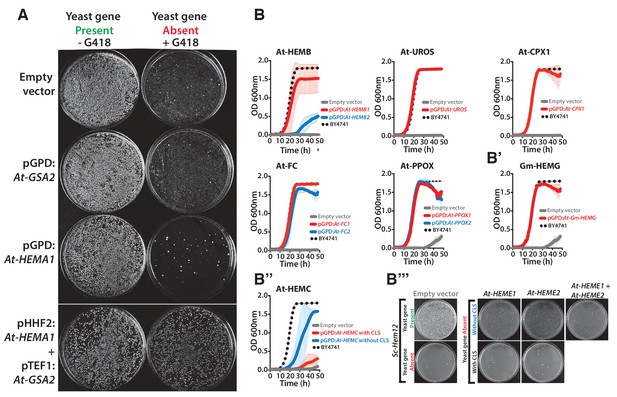
Heme biosynthesis genes from Arabidopsis thaliana and Glycine max generally efficiently replace their counterparts in yeast, except in the case of ΔSc-Hem12.
(A) Expression of heme pathway genes from Arabidopsis thaliana, At-HEMA1 or At-GSA2, individually cannot complement the lethal growth defect of the deletion of Sc-hem1 gene in yeast. Co-expression of At-HEMA1 and At-GSA2 rescued the growth defect of Sc-hem1 gene deletion in yeast. (B) Haploid yeast gene deletion strains carrying plasmids expressing functionally replacing Arabidopsis (red or blue solid-lines) and (B’) Glycine max (Gm-HEMG) heme pathway genes (red solid-line) generally exhibit comparable growth rates to the wild type parental yeast strain BY4741 (black dotted-line) as grown in magic marker liquid medium in the presence of G418 (200 μg/ml). (B’’) Native At-HEMC with chloroplast localization signal (CLS) showed poor replaceability in yeast (red solid-line). Removal of the CLS from At-HEMC allowed efficient rescue of the corresponding yeast gene deletion, ΔSc-Hem3 (blue solid-line). (B’’’) However, neither the expression of Arabidopsis proteins At-HEME1 or At-HEME2 (with or without CLS) alone nor their co-expression could functionally rescue the corresponding yeast gene deletion, ΔSc-Hem12. Wild type BY4741 haploid strain is plotted for comparison (black dotted-line). Strains carrying empty vector were used as controls (grey solid-line). Mean and standard deviation plotted with N = 3.
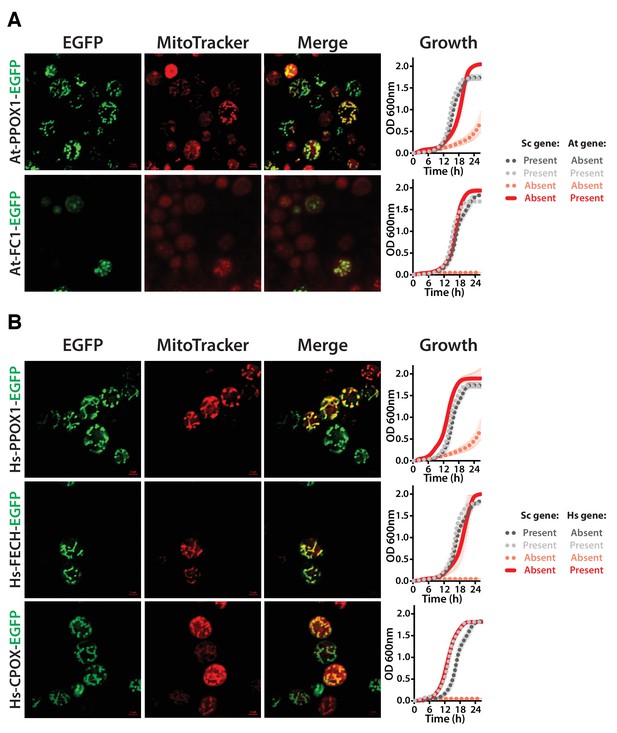
Heme biosynthesis enzymes normally localized to plant chloroplasts or human mitochondria localize to the mitochondria when expressed in yeast.
(A) EGFP-tagged penultimate At-PPOX1-EGFP and ultimate At-FC1-EGFP proteins localize to mitochondria in yeast. Green fluorescence proteins co-localized with Mitotracker red-stained mitochondria. In certain cases, At-FC1-EGFP formed aggregates. Expression of EGFP-tagged plant genes, At-PPOX1-EGFP and At-FC1-EGFP (red solid-line), efficiently rescue the growth defect of the corresponding yeast gene deletions (pink dotted-line). The over-expression of the tagged proteins is not toxic to the wild type yeast strain (grey dotted-line). The growth rescue by plant genes is as efficient as the wild type BY4741 yeast strain (black dotted-line). Mean and standard deviation plotted with N = 3. (B) The EGFP-tagged last three heme pathway genes from humans localize to mitochondria in yeast. The green fluorescence co-localized with the Mitotracker red-stained mitochondria in yeast. Expression of EGFP-tagged human genes, Hs-PPOX-EGFP, Hs-FECH-EGFP and Hs-CPOX-EGFP (red solid-line), efficiently rescue the growth defect of the corresponding yeast gene deletions (pink dotted-line). The over-expression of the tagged proteins is not toxic to the wild type yeast strain (grey dotted-line). The growth rescue by the human genes is as efficient as the wild type BY4741 yeast strain (black dotted-line). Mean and standard deviation plotted with N = 3.
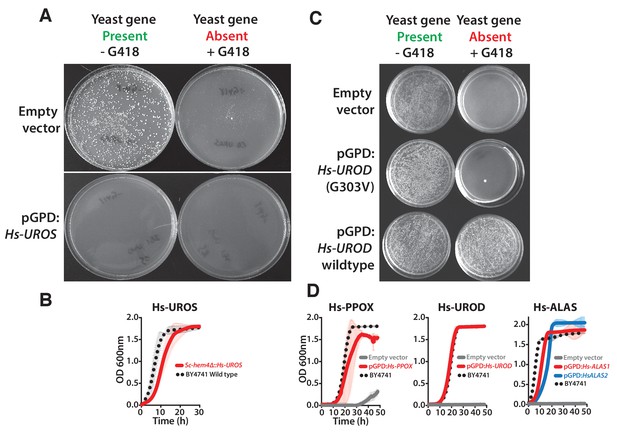
Human heme biosynthesis genes efficiently replace their yeast counterparts.
Functional replacement of human genes in yeast. (A) Expression of Hs-UROS in Sc-hem4 heterozygous diploid deletion yeast strain resulted in toxicity post-sporulation as seen by the lack of growth on either magic marker agar medium with (yeast gene present) or without G418 (yeast gene absent). (B) This toxicity was relieved by replacing the human Hs-UROS at the native yeast locus. Growth curve of the humanized yeast Sc-hem4Δ::Hs-UROS strain (red-solid line) showed comparable growth to the wild type yeast BY4741 (black dotted-line). (C) Expression of human Hs-UROD (a human orfeome clone with G303V mutation) in Sc-hem12 heterozygous diploid deletion yeast strain did not complement the growth defect of the yeast gene as shown by plating the post sporulation mix on magic marker medium with or without G418. Reverting the sequence to the wild type Hs-UROD gene resulted in efficient rescue of the growth defect of the corresponding yeast gene. (D) Expression of human genes, Hs-PPOX, Hs-UROD, Hs-ALAS1 (red solid-line) and Hs-ALAS2 (blue solid-line), efficiently rescue the growth defect of the corresponding yeast gene deletions (grey solid-line), Sc-hem14 and Sc-hem1, respectively. The rescue was largely comparable to the wild type BY4741 yeast strain (black dotted-line). Strains carrying empty vector were used as controls (grey solid-line). Mean and standard deviation plotted with N = 3.
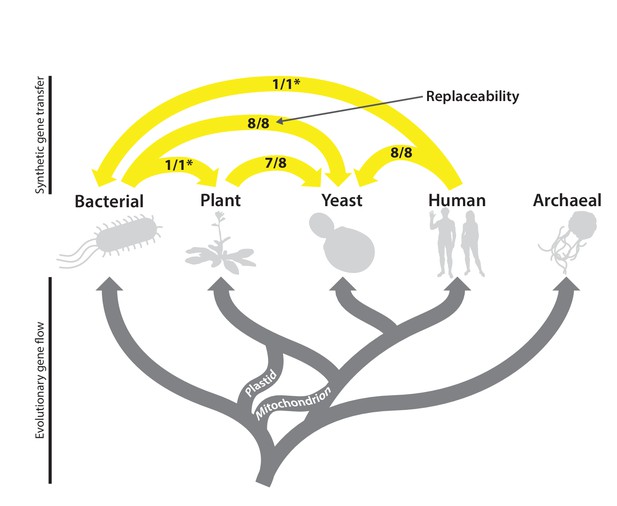
The complex evolutionary history of the heme biosynthesis pathway is reflected in high replaceability across species.
In eukaryotes, heme biosynthesis enzymes have been replaced historically by endosymbiosis events from bacteria, leading to higher similarity across these lineages, while the archaeal pathway appears to be more divergent (Storbeck et al., 2010). Following the endosymbiosis of the cyanobacterial chloroplast, plants adopted most of the chloroplast-derived heme biosynthesis genes, losing many ancestral eukaryotic heme pathway genes (Oborník and Green, 2005). Yeast and humans both retain the predicted ancestral eukaryotic heme biosynthesis pathway. While enzymatic steps are mostly shared between yeast, plants, bacteria, and humans, localization of individual proteins differs substantially between species. Asterisks indicate results curated from literature.
Additional files
-
Supplementary file 1
Detailed results of complementation assays.
Rows are ortholog pairs. Columns A-E list several alternative gene IDs for each organism. ‘Assay location’ is the location of that complementation assay in the plate images shown in Figure 1—figure supplement 1. The following columns list the results of the specific assay type: ‘MM’ refers to the heterozygous diploid assay (Magic Marker). ‘TS’ refers to the temperature-sensitive allele assay. ‘With MLS’ are assays re-done with the yeast mitochondrial localization sequence as described in the test. ‘With ATG’ are assays re-done with an ATG start codon substituted for the E. coli genes non-canonical start codon. ‘Plasmid dependence’ lists the results of 5’ FOA screening of complementing clones to confirm that the E. coli gene-containing plasmid is present. ‘Final status’ and ‘Preliminary status’ refer to the complementation status of the given ortholog pair after (Final) or before (Preliminary) accounting for MLS, ATG, or plasmid dependence assays.
- https://doi.org/10.7554/eLife.25093.022
-
Supplementary file 2
Data used to calculate predictive features.
The first sheet displays all data for each property used in the study to determine predictive properties. The first several columns list IDs for the two organisms as well as the final complementation results. The following columns include all data for each feature. See Materials and Methods for detailed descriptions of each property. The second sheet lists the calculated AUCs for each feature, and the results of the shuffling procedure for each.
- https://doi.org/10.7554/eLife.25093.023
-
Supplementary file 3
Primers used in this study.
A brief description of each primer's use is included on each separate sheet of the file. For additional information, see Materials and methods for the relevant section.
- https://doi.org/10.7554/eLife.25093.024





