Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration
Figures
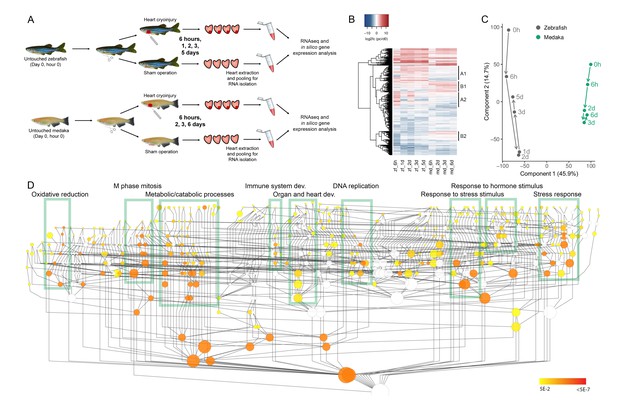
Comparative transcriptomic analyses between zebrafish and medaka after cardiac injury.
(A) Experimental design. Untouched, sham-operated, and cryoinjured zebrafish and medaka ventricles were collected at 6 hr, 1 d (zebrafish only), 2 d, 3 d, and 5 d (zebrafish only)/6 d (medaka only) post-surgery. Four hearts per time point were pooled, RNA was extracted and subjected to sequencing. (B) Global heatmap depicting expression values of differentially expressed genes vs untouched (0 hr). Hierarchical clustering of genes with an absolute FC of log2 >2 in at least one sample is shown in a heatmap. FC was calculated by comparing to the respective 0 hr expression value. Red color indicates higher expression in the respective sample listed at the bottom of each column, blue color indicates higher expression in the 0 hr sample of the respective organism (medaka or zebrafish). Clusters containing differentially regulated genes between zebrafish and medaka are indicated by A1, A2, B1, and B2. (C) PCA with dimension 1 and 2 of normalized counts. PCA was performed on the normalized counts of untouched and cryoinjured fish, and the first two dimensions are visualized capturing ~60% of the variability of the dataset. (D) Gene Ontology (GO) enrichment of biological processes. Schematic illustration of a GO tree based on biological processes. Each GO term is indicated as a circle whose radius reflects the number of proteins assigned to it. Coloring indicates Benjamini Hochberg corrected p-values<0.05 of hypergeometric tests for overrepresentation/underrepresentation as given in the color scale on the right. The test was performed against the whole annotation reference set. Boxes with respective description mark subtrees of the GO graph that are highly connected and significantly enriched. The given description reflects one of the most enriched terms in the subtree.
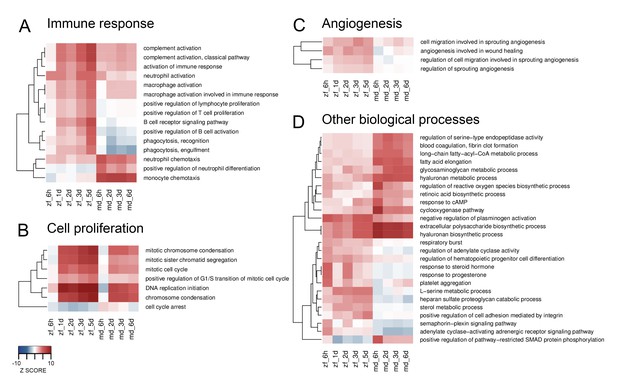
Heatmaps of differentially enriched GO terms from SLEA.
GO terms from SLEA analysis of cryoinjured vs. untouched hearts related to immune response (A), cell proliferation (B), angiogenesis (C) and other biological processes (D), were selected and respective Z-scores visualized. Color scales were matched for all four heatmaps, ranking from –10 for underrepresentation to +10 for overrepresentation.
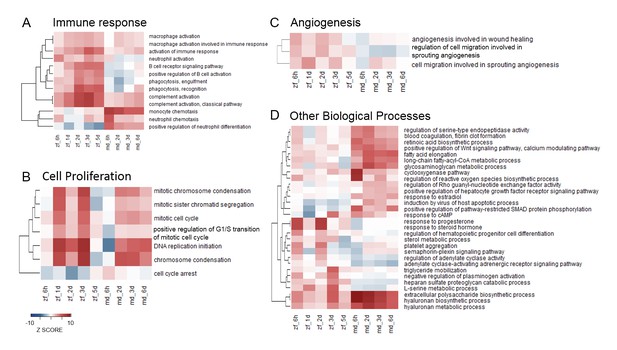
Heatmaps of enriched GO terms from SLEA.
GO terms from SLEA analysis of sham-operated vs. untouched hearts related to immune response (A), cell proliferation (B), angiogenesis (C) and other biological processes (D), were selected and respective Z-scores visualized. Color scales were matched for all four heatmaps, ranking from –10 for underrepresentation to +10 for overrepresentation.
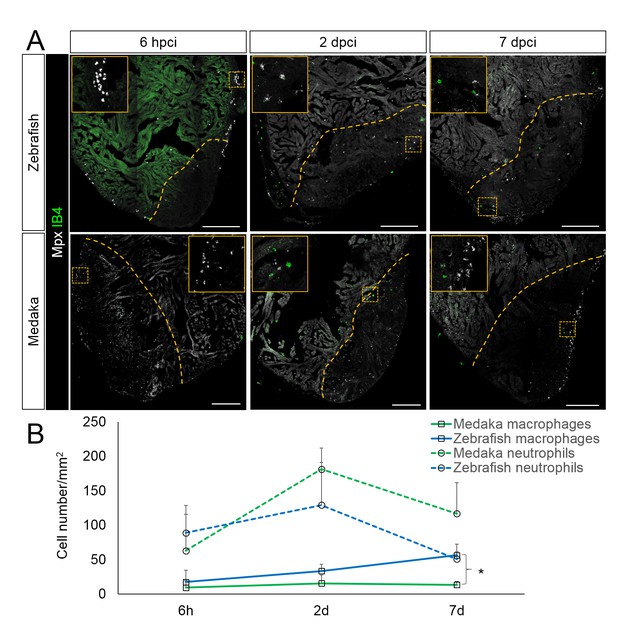
Immune cells dynamics in cryoinjured hearts.
(A) Zebrafish and medaka heart sections at 6 hpci, 2 dpci and 7 dpci were stained with isolectin-B4 (IB4) for macrophages, and Mpx antibody for neutrophils. Positive cells, both in the injured area itself and within 100 μm of the injured area, were quantified (B; n = 3). Dotted lines delineate the injured area; scale bars, 200 μm. Macrophage numbers in medaka were always lower than those in zebrafish and this difference was especially pronounced at 7 pd. Neutrophil numbers appeared similar at 6 hpci, were higher in medaka at 2 dpci, and their clearance was delayed in medaka compared to zebrafish.
-
Figure 3—source data 1
Quantification of immune cells in cryoinjured hearts.
- https://doi.org/10.7554/eLife.25605.006
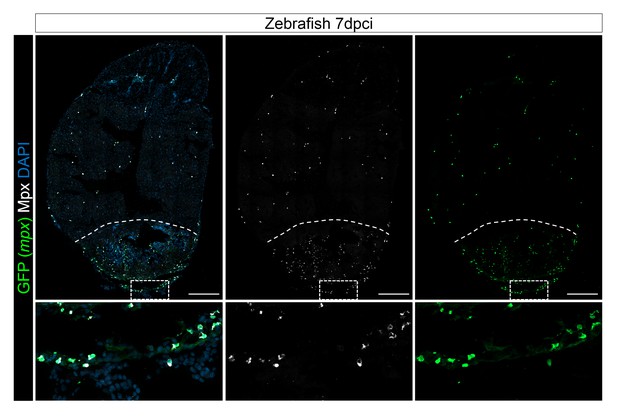
Mpx antibody specifically stains mpx:GFP-expressing cells.
Injured hearts from adult mpx:GFP zebrafish were collected at 7 dpci, and cryosections stained with GFP and Mpx antibodies to examine neutrophils. Dotted lines delineate the injured area; scale bars, 200 μm. Percentage of Mpx+;GFP+ cells/GFP+ cells was quantified. 88.18 ± 0.57% of GFP+ cells were Mpx+, and we did not observe any Mpx-;GFP+ cells (Figure 3—figure supplement 1—source data 1).
-
Figure 3—figure supplement 1—source data 1
Quantification of Mpx+ and mpx:GFP+ cells in zebrafish hearts following cardiac injury.
- https://doi.org/10.7554/eLife.25605.008
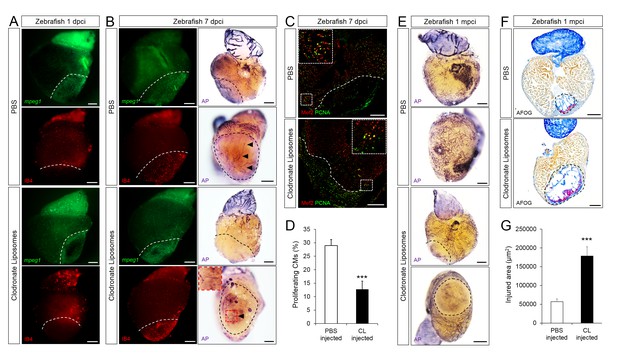
Delayed macrophage recruitment in zebrafish compromises neovascularization, CM proliferation and scar resolution.
Adult zebrafish were injected with PBS or clodronate liposomes 1 day before injury. Hearts were collected at 1 dpci (A), 7 dpci (B and C) and 1 mpci (E–G) to examine macrophages by mpeg1:EGFP expression and IB4 staining (A and B), neovascularization by alkaline phosphatase (AP) staining (B and E, a side view as well as an apex view are shown), CM proliferation by PCNA/Mef2 immunostaining (C), and scar resolution by Acid Fuchsin Orange G (AFOG) staining (F). Mef2 and PCNA double positive CMs within 200 μm of the injured area were quantified in (D). Sections with the largest injured area/scar for each 1 mpci heart (delineated by black dotted lines) were stained with AFOG and quantified in G (n > 5, shown in Figure 4—figure supplement 2). Dotted lines delineate the injured area, arrowheads point to vessels, asterisks mark the initial injury site; scale bars, 200 μm. N ≥ 3 for each treatment and time point. Clodronate injections diminished macrophage recruitment at 1 dpci, and macrophage numbers recovered to control levels at 7 dpci (A and B, quantification in Figure 4—figure supplement 1 and Figure 4—figure supplement 1—source data 1). Delayed macrophage recruitment compromised neovascularization (B and E), and CM proliferation (C and D), and resulted in delayed scar resolution (F and G).
-
Figure 4—source data 1
Quantification of proliferating CMs in control and CL-exposed zebrafish hearts following cardiac injury.
- https://doi.org/10.7554/eLife.25605.010
-
Figure 4—source data 2
Quantification of scar areas in control and CL-exposed zebrafish hearts following cardiac injury.
- https://doi.org/10.7554/eLife.25605.011
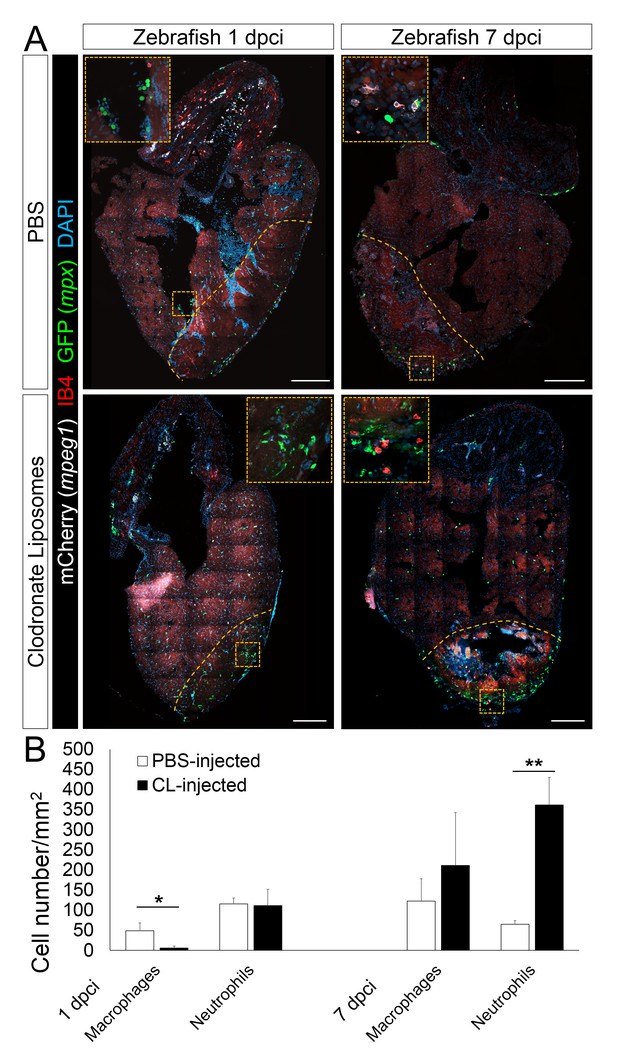
Delayed macrophage recruitment in zebrafish by Clodronate liposome pre-depletion.
Adult mpeg1.4:mCherry-F;mpx:GFP zebrafish were injected with PBS or clodronate liposomes 1 day before injury. Hearts were collected at 7 dpci, and cryosections stained with GFP antibody to examine neutrophils, and mCherry antibody and IB4 to examine macrophages. Positive cells, both in the injured area itself and within 100 μm of the injured area, were quantified (B; n ≥ 3). Dotted lines delineate the injured area; scale bars, 200 μm. At 1 dpci, macrophage numbers in CL-exposed hearts were significantly lower than in controls, while neutrophils numbers were similar. At 7 dpci, neutrophil numbers were significantly higher in CL-exposed hearts, while macrophage numbers were not significantly different.
-
Figure 4—figure supplement 1—source data 1
Quantification of macrophage and neutrophil numbers in control and CL-exposed zebrafish hearts following cardiac injury.
- https://doi.org/10.7554/eLife.25605.013
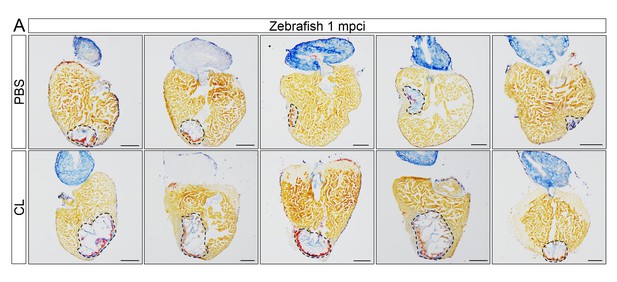
Delayed macrophage recruitment in zebrafish compromises scar resolution.
Adult zebrafish were injected with PBS or clodronate liposomes 1 day before injury. Hearts were collected at 1 mpci, and cryosections were stained with AFOG. Healthy muscle stained in orange, fibrin in red, and collagen in blue. Sections with the largest scar area for each heart (delineated by dotted lines) were selected and quantified (Figure 4G, n = 5). Scale bars, 200 μm. Scar tissue in CL-exposed hearts was significantly larger than in controls at 1 mpci.
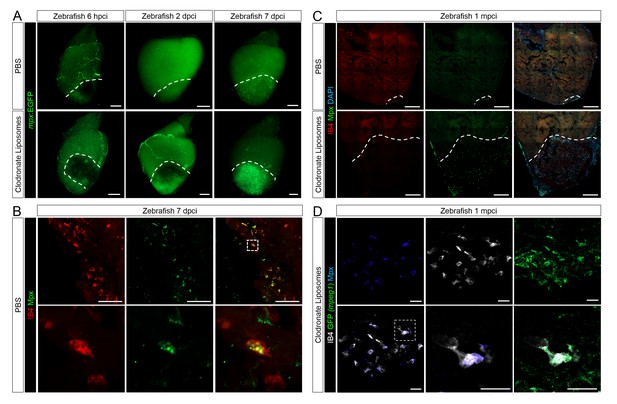
Neutrophil clearance is delayed after macrophage pre-depletion.
Adult zebrafish were injected with PBS or clodronate liposomes 1 day before injury. Hearts were collected at 6 hpci (A), 2 dpci (A), 7 dpci (A and B), and 1 mpci (C and D) to examine macrophages by both IB4 staining and mpeg1:EGFP expression, and neutrophils by immunostaining for both Mpx and mpx:GFP expression. (A) Neutrophil dynamics at 6 hpci, 2 and 7 dpci. (B) Neutrophil clearance by macrophage phagocytosis was observed in PBS-exposed hearts at 7 dpci. (C) Neutrophils remained in the injured area of CL-exposed hearts at 1 mpci. (D) Neutrophil clearance by IB4- and mpeg1:EGFP-double positive macrophages was observed in CL-exposed hearts at 1 mpci. Dotted lines delineate injured area; scale bars, 200 μm (A), 50 μm (B), 100 μm (C), and 20 μm (D).
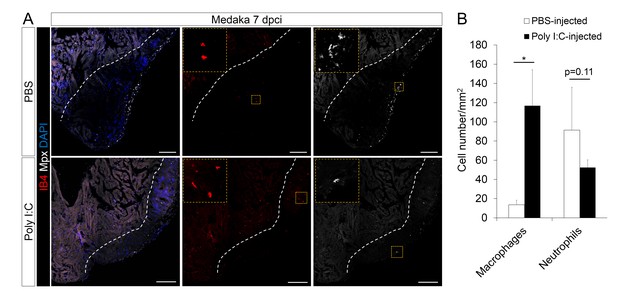
Poly I:C injection in medaka promotes macrophage recruitment and neutrophil clearance following cardiac injury.
(A) Medaka heart sections at 7 dpci were stained with IB4 for macrophages and Mpx antibody for neutrophils. Positive cells, both in the injured area itself and within 100 μm of the injured area, were quantified (B; n = 3). Dotted lines delineate the injured area; scale bars, 100 μm. Poly I:C injection significantly promoted macrophage recruitment and neutrophil clearance in cryoinjured medaka hearts at 7 dpci.
-
Figure 6—source data 1
Quantification of macrophage and neutrophil numbers in control and poly I:C-exposed medaka hearts following cardiac injury.
- https://doi.org/10.7554/eLife.25605.017
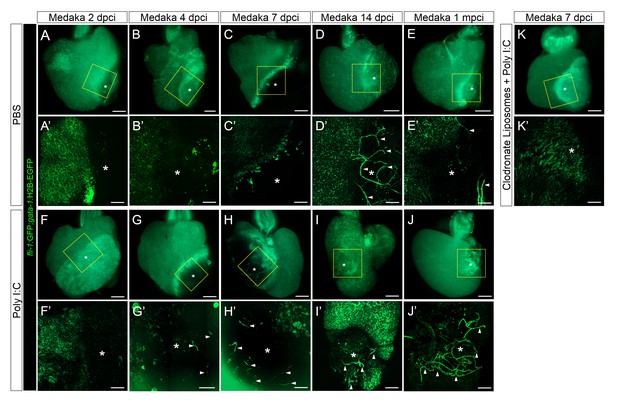
Poly I:C injection in medaka promotes neovascularization in a macrophage-dependent manner following cardiac injury.
Adult fli::GFP;gata1::GFP medaka were injected with PBS (A–E) or poly I:C (F–J) immediately after injury, or with clodronate liposomes at 1 day before injury and poly I:C immediately after injury (K). Hearts were collected at 2 (A and F), 4 (B and G), 7 (C, H, and K), and 14 (D and I) dpci, as well as 1 mpci (E and J), and imaged with a stereo (A–K) or confocal (A’-K’, boxed areas in A-K) microscope for detailed examination. Asterisks mark the injured area, and arrowheads point to new vessel-like structures. Initiation of vessel formation was observed at earlier time points (4 and 7 dpci) in poly I:C-exposed hearts compared to control hearts, and these vessels were maintained at least until 1 mpci (n > 3). In contrast, in control hearts, new vessels were observed only at 14 dpci, and largely disappeared by 1 mpci. In addition, vessel formation promoted by poly I:C injections was blocked by CL pre-injections.
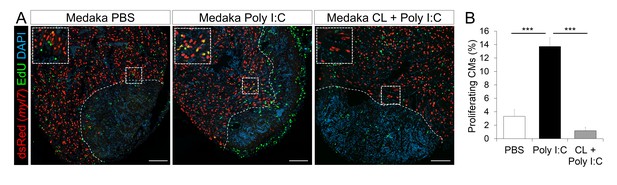
Poly I:C injection in medaka promotes CM proliferation in a macrophage-dependent manner following cardiac injury.
Adult zfmlc2 5.1 k:DsRED2-nuc medaka were injected with PBS or poly I:C immediately after injury, or with clodronate liposomes at 1 day before injury and poly I:C immediately after injury. Hearts were collected at 7 dpci, and CM proliferation was examined by EdU labeling and immunostaining for DsRED expression; dotted lines delineate the injured area. EdU positive CMs within 200 μm of the injured area were quantified and shown (B; n ≥ 3). Scale bars, 100 μm. The percentage of proliferating CMs was significantly higher in poly I:C-exposed hearts compared to both PBS- and CL + poly I:C-exposed hearts.
-
Figure 8—source data 1
Quantification of proliferating CMs in medaka hearts following cardiac injury.
- https://doi.org/10.7554/eLife.25605.020
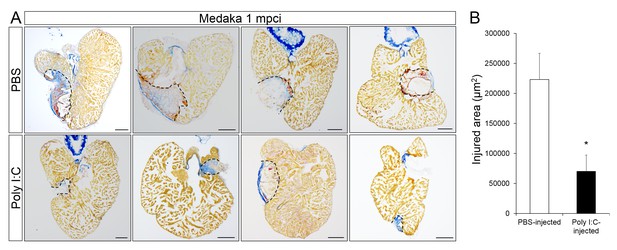
Poly I:C injection in medaka promotes scar resolution following cardiac injury.
Adult fli::GFP;gata1::GFP medaka were injected with PBS or poly I:C immediately after injury. Hearts were collected at 1 mpci, and scar composition and resolution were examined by AFOG staining. (A) Healthy muscle stained in orange, fibrin in red, and collagen in blue. Sections with the largest scar area for each heart (delineated by dotted lines) were selected and quantified (B; n ≥ 4). Scale bars, 200 μm. Scar tissue in poly I:C-exposed hearts was significantly smaller than in controls at 1 mpci.
-
Figure 9—source data 1
Quantification of scar areas in medaka hearts following cardiac injury.
- https://doi.org/10.7554/eLife.25605.022
Additional files
-
Supplementary file 1
Normalized orthologous genes expression in zebrafish and medaka post cardiac injury.
- https://doi.org/10.7554/eLife.25605.023
-
Supplementary file 2
List of biological processes revealed by Sample Level Enrichment Analysis.
- https://doi.org/10.7554/eLife.25605.024
-
Supplementary file 3
Top canonical pathways and upstream regulators predicted by Ingenuity Pathway Analysis.
(Z-zebrafish, M-medaka, H-hours post injury, D-days post injury.)
- https://doi.org/10.7554/eLife.25605.025
-
Supplementary file 4
List of canonical pathways and upstream regulators predicted by Ingenuity Pathway Analysis.
- https://doi.org/10.7554/eLife.25605.026





