Elastic force restricts growth of the murine utricle
Figures
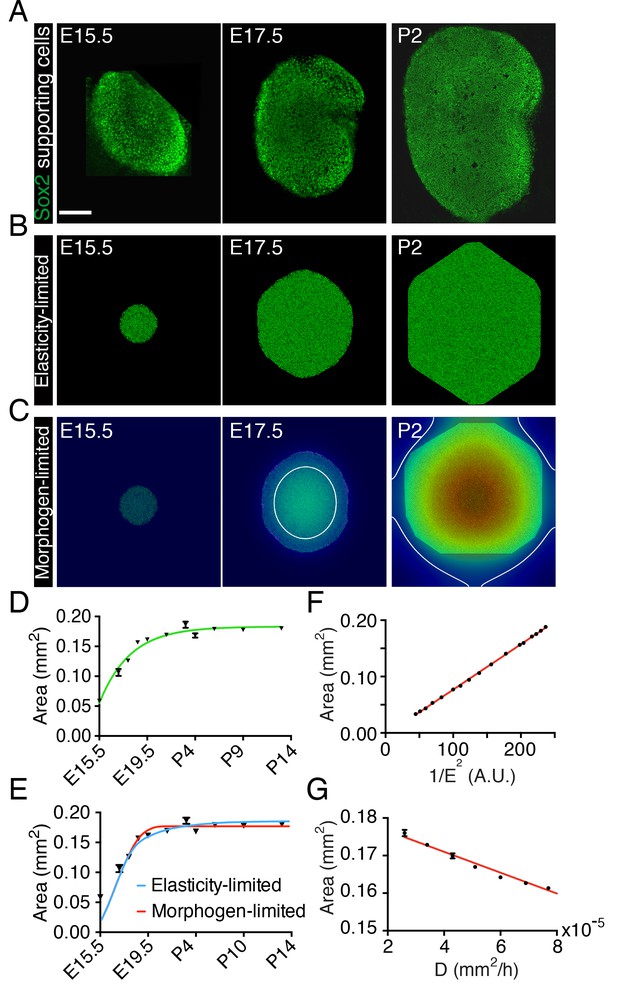
Size control in utricular development.
(A) Immunolabeling of supporting cells with anti-Sox2 (green) demonstrates the size of the utricular sensory epithelia of mice at E15.5, E17.5, and P2. The scale bar represents 100 μm. (B) Simulations of the elasticity-limited model portray sensory epithelia at the same developmental stages (Source code 1; Table 1). (C) In simulations of the morphogen-limited model, the concentration of the diffusible morphogen is shown on a spectral scale from blue as zero to red as the maximum, 0.2 pM. The white curve bounds the region in which a concentration exceeding the threshold value = 0.04 pM inhibits cellular proliferation (Source code 2; Table 1). (D) The measured areas of sensory epithelia at different developmental stages (black triangles; means ± SEMs; Figure 1—source data 1) are fit by the von Bertalanffy equation (green curve; = 1.8·10−7 ± 5.8·10−9 m2, = 3.6·10−6 ± 5.8·10−7 s−1, = −1.32·10−5 ± 3.7·10−6 s−1, R2 = 0.94). (E) The same data are portrayed with fits of the elasticity-limited model (blue curve, R2 = 0.74) and morphogen-limited model (red curve, R2 = 0.62). Each result represents the average of five realizations; the SEM is typically around 1% (Figure 1—source datas 3,5). (F) Final areas of utricular sensory epithelia from simulations of the elasticity-limited model (means ± SEMs, N = 3) for different values of the Young’s modulus of the elastic boundary E. The areas are proportional to 1/E2 (black dots) and fit the linear relation Y = 0.000804·X - 0.00361 (red line, R2 = 0.99; Figure 1—source data 2). (G) Final areas of utricular sensory epithelia from simulations of the morphogen-limited model (means ± SEMs, N = 3) are linearly related to the diffusion coefficient (black dots) by the relation Y = −278·X - 0.182 (red line, R2 = 0.98; Figure 1—source data 4).
-
Figure 1—source data 1
The areas of utricular sensory epithelia measured at different developmental stages.
- https://doi.org/10.7554/eLife.25681.003
-
Figure 1—source data 2
Final areas of utricular sensory epithelia from simulations of the elasticity-limited model for different values of the Young’s modulus of the elastic boundary .
- https://doi.org/10.7554/eLife.25681.004
-
Figure 1—source data 3
Fits of the elasticity-limited model to the areas of utricular sensory epithelia measured at different developmental stages.
- https://doi.org/10.7554/eLife.25681.005
-
Figure 1—source data 4
Final areas of utricular sensory epithelia from simulations of the morphogen-limited model for different values of the diffusion coefficient .
- https://doi.org/10.7554/eLife.25681.006
-
Figure 1—source data 5
Fits of the morphogen-limited model to the areas of utricular sensory epithelia measured at different developmental stages.
- https://doi.org/10.7554/eLife.25681.007
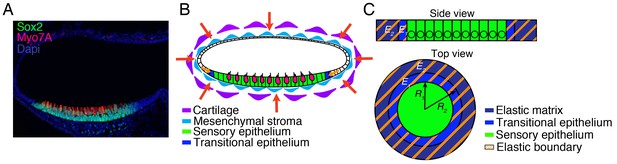
Geometry of the utricle and simplification to a mathematical model.
(A) In a transverse section of a utricle at E17.5, Sox2-positive cells (green) form the sensory epithelium that sits at the bottom of the fluid-filled organ. Myo7A (red) labels hair cells. DAPI (blue) stains the nuclei of all the cells in the utricle and demonstrates the presence of Sox2-neganive non-sensory cells that are contingent with the sensory epithelium and are lining the rest of the utricle. The mesenchyme surrounding the utricle is also visualized by DAPI staining. (B) In a schematic representation, the utricle is surrounded by mesenchymal cells (light blue) that attach it to the surrounding cartilage (purple). We term both these tissues the elastic matrix. This matrix exerts compressive elastic forces that oppose the expansion of the utricle (red arrows), These forces are transmitted tangentially to the inner layer of the ellipsoidal chamber and act on the transitional non-sensory epithelium (blue), which then opposes the expansion of the sensory epithelium (green and magenta). The effective elastic force is the sum of that owing to the transitional epithelium and the tangential force applied by the elastic matrix (orange arrows). (C) We simplify the treatment of the model by considering the utricle as a two-dimensional structure. The sensory epithelium (green) forms a surface that is surrounded by a band of translational epithelium of inner radius and outer radius and with a Young’s modulus (blue). This epithelium is in turn bounded by an elastic matrix of inner radius and Young’s modulus (purple). The transitional epithelium and elastic matrix can be treated as one effective material of Young’s modulus , which we term the surrounding elastic band (orange fringes).
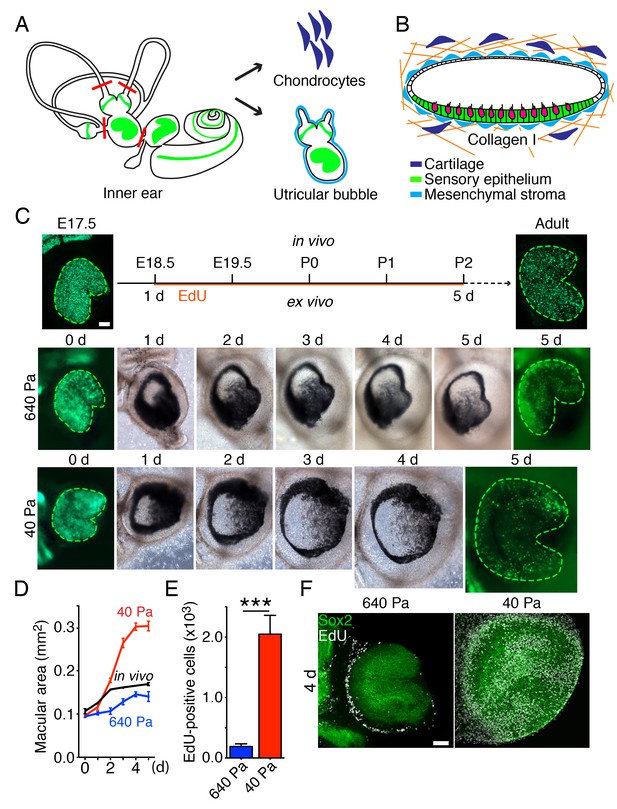
Ex vivo utricular culture system.
(A) A schematic drawing portrays the sensory epithelia (green) of the six receptor organs of the murine inner ear. The red lines delineate the cuts introduced during the dissection of a utricular bubble. The mesenchymal stroma that normally surrounds the organ is shown in light blue; the chondrocytes that surround the inner ear are shown in dark blue. (B) In a diagram of a section through a utricular bubble, hair cells are shown in magenta, supporting cells in green, non-sensory cells in white, and mesenchyme in light blue. The bubble is embedded in a gel of collagen I (orange) containing chondrocytes (dark blue). (C) A sequence of surface views portrays the development of two utricular bubbles from E17.5 Atoh-nGFP mice over a period of 5 d. The sensory epithelia are marked in the first and final images by GFP expression (green; delineated by green dashed line). In a gel of 640 Pa stiffness (middle panels), the utricular sensory epithelium reaches the normal adult size (in vivo; upper panels) by 5 d, corresponding to developmental stage P2. In a collagen gel of 40 Pa stiffness (lower panels) the area of the sensory epithelium almost doubles by 5 d. The scale bar represents 50 μm. (D) Areal measurements, represented as means ± SEMs, demonstrate that the utricular sensory epithelium in a 640 Pa gel develops at a rate similar to that of the epithelium in vivo. By 5 d in culture the area of the sensory epithelium is similar to that of an adult utricle (p<0.05, N = 5 for both; Figure 2—source data 1). In a 40 Pa gel, the area of the sensory epithelium expands significantly faster and reaches 180% of the control value by 5 d (p<0.0001, N = 4 for 40 Pa; N = 5 for in vivo measurements; Figure 2—source data 1). (E) After 4 d in culture, quantification of EdU-positive cells in 0.1 mm2 of the utricular sensory epithelium, represented as means ± SEMs, demonstrates a tenfold increase in the number of EdU-positive cells in a 40 Pa gel as compared with a 640 Pa gel. This difference is significant (p<0.001, N = 3 for both; Figure 2—source data 2) (F) A surface view of utricular bubbles after 4 d in culture demonstrates that the expansion of the macula in a 40 Pa gel stems from the re-entry of Sox2-positive supporting cells (green) into the mitotic cycle throughout the sensory epithelium, as demonstrated by the incorporation of EdU (white). In the identical preparation of the utricular bubble cultured in a 640 Pa gel, supporting-cell proliferation is limited to the organ’s periphery. The scale bar represents 50 μm.
-
Figure 2—source data 1
The area of the sensory epithelium of the utricles cultured in 40 Pa and 640 Pa gels for 1–5 d.
- https://doi.org/10.7554/eLife.25681.013
-
Figure 2—source data 2
The number of EdU-positive cells in 0.1 mm2 of the utricular sensory epithelium in 40 Pa and 640 Pa gels after 4 d in culture.
- https://doi.org/10.7554/eLife.25681.014
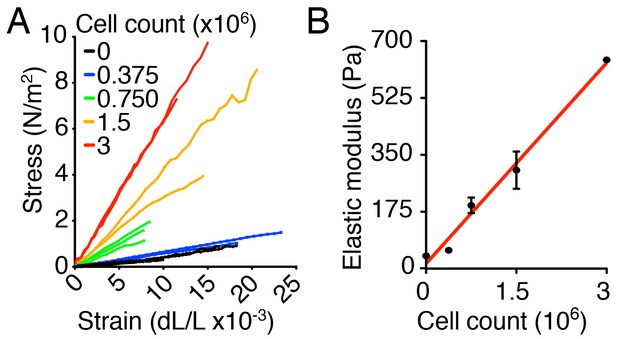
Characterization of the ex vivo utricular culture system.
(A) The slopes of the stress-strain relations for individual gels, corresponding to the elastic moduli of the materials, rise as the number of chondrocytes increases from zero to 3·106. (B) The stiffness of the collagen gel increases linearly with the number of chondrocytes. The relation is fit by the linear function = 206· +15, in which is the elastic modulus and is the number of chondrocytes (R = 0.99, p<0.001).
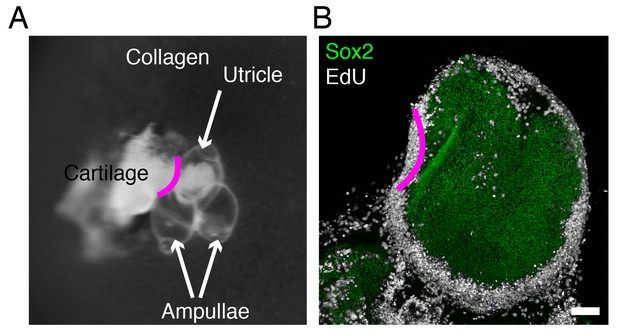
Chondrocyte-secreted factors do not affect supporting-cell proliferation.
(A) In a brightfield image of a utricular bubble culture established at E17.5, the utricle and ampullae semicircular canals are labeled. A piece of cartilage was dissected from the same E17.5 inner ear and positioned near the edge of the utricle, indicated by the magenta line. (B) In a utricular bubble preparation after 24 hr in culture, immunolabeling for Sox2 (green) reveals the size and position of the sensory epithelia of the utricle; EdU (white) demonstrated the distribution of proliferating cells adjacent to (pink line) and distant from the cartilage. The scale bar represents 100 μm.
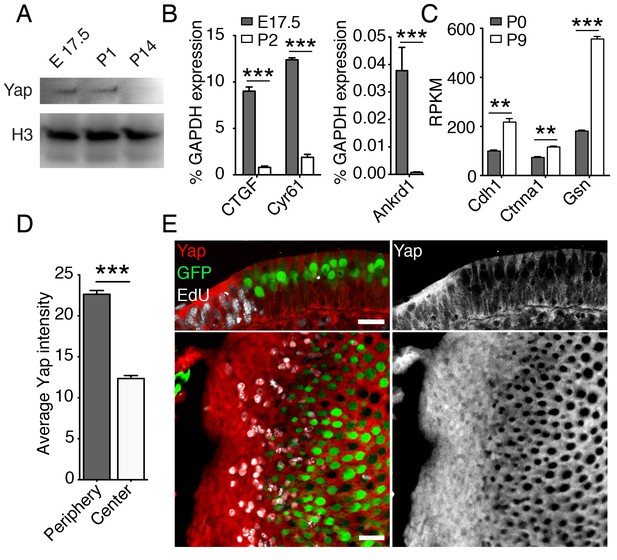
Yap signaling during utricular development.
(A) Western blot analysis of the total protein extracted from normally developing murine utricles demonstrates a progressive decrease in the level of Yap protein. (B) qPCR analysis of normally developing murine utricles reveals significant decreases in the expression of the Yap target genes Ctgf, Cyr61, and Ankrd1 between E17.5 and P2 (means ± SEMs; p<0.001 for each; N = 6 for E17.5; and N = 9 for P2; Figure 3—source data 1). (C) RNA sequencing from normally developing murine utricles reveals a progressive increase in the expression of genes encoding inhibitors of Yap tranlocation to the nucleus—Cdh1, Ctnna1, and Gsn—between P0 and P9 (means ± SEMs; p<0.005 for Cdh1, p<0.005 for Ctnna1, and p<0.0001 for Gsn; N = 3 for each; Figure 3—source data 2). (D) The mean fluorescence intensity of antibody labeling for Yap is significantly higher at the periphery than near the center of the utricle (value range 0–85; means ± SEMs; p<0.0001; N = 4 for each; Figure 3—source data 3) (E) In cross-sections (top panels) of the peripheral utricular sensory epithelium of E17.5 Atoh-nGFP mice 4 hr after EdU treatment, antibody labeling demonstrates Yap protein (red at left; white at right) in actively proliferating cells labeled with EdU (white at left). Hair cells (green) do not express Yap. In a different specimen this pattern of labeling is demonstrated in surface views (bottom panels). The scale bar represents 25 μm.
-
Figure 3—source data 1
qPCR analysis of the expression of the Yap target genes Ctgf, Cyr61, and Ankrd1 at E17.5 and P2.
- https://doi.org/10.7554/eLife.25681.018
-
Figure 3—source data 2
RNA sequencing of the expression of genes encoding inhibitors of Yap tranlocation to the nucleus—Cdh1, Ctnna1, and Gsn—at P0 and P9.
- https://doi.org/10.7554/eLife.25681.019
-
Figure 3—source data 3
The mean fluorescence intensity of antibody labeling for Yap at the periphery and near the center of the E17.5 utricle.
- https://doi.org/10.7554/eLife.25681.020
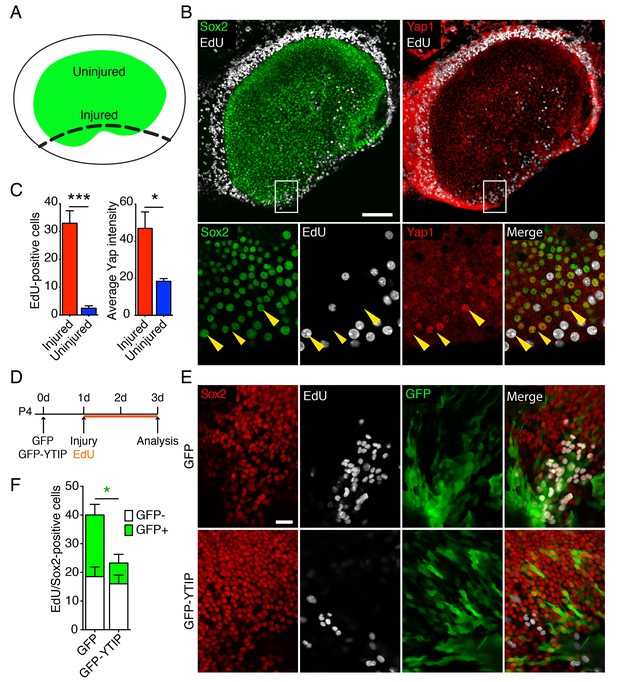
Yap distribution during utricular regeneration after injury in vitro.
(A) A schematic drawing portrays the sensory epithelium (green) bounded by non-sensory epithelial cells (white). The black dashed line delineates the lesion. (B) In P4 utricles cultured for 48 hr in vitro, Sox2-positive supporting cells (green) at the injury site, as well as the transitional non-sensory epithelial cells that bound the sensory epithelium, express high levels of Yap protein (red). As shown by the incorporation of EdU (white), the same cells re-enter the cell cycle. Yap nuclear translocation can be seen in individual Sox2-positive supporting cells at the injury site (magnified 4X in lower panels; yellow arrowheads). The scale bar represents 100 μm. (C) The number of EdU-positive cells in the injured areas of utricular sensory epithelia rises significantly in comparison to noninjured areas of the same epithelia (cells counted over 0.01 mm2; means ± SEMs; p<0.001, N = 4 for injured areas and N = 8 for noninjured areas; Figure 4—source data 1). The amount of proliferation correlates with the average intensity of Yap immunolabeling, which is significantly higher in injured areas (value range 0–85; means ± SEMs; p<0.05, N = 6 for injured areas and N = 5 for noninjured areas; Figure 4—source data 2). (D) A schematic representation of the lesioning experiment demonstrates the time at which the utricles are transfected with GFP and GFP-YTIP virus, the time of injury, and the duration of EdU labeling. (E) In P4 utricles cultured for 3 d in vitro, Sox2-positive supporting cells (red) at the injury site enter the cell cycle, as shown by EdU incorporation (white). Cells expressing GFP and GFP-YTIP are shown in green. The scale bar represents 25 μm. (F) Although the number of GFP-negative proliferating supporting cells remains unchanged between the conditions (white bars; cells counted over 0.01 mm2; means ± SEMs; p>0.05, N = 4 for each; Figure 4—source data 3), the number of GFP-positive proliferating supporting cells is significantly reduced at the injury site of the utricles expressing GFP-YTIP as compared to cntrols expressing GFPs (green bars; cells counted over 0.01 mm2; means ± SEMs; p<0.025, N = 4 for each; Figure 4—source data 3).
-
Figure 4—source data 1
The number of EdU-positive cells in the injured and noninjured areas of P4 utricular sensory epithelium after 24 hr in culture.
- https://doi.org/10.7554/eLife.25681.023
-
Figure 4—source data 2
The mean fluorescence intensity of antibody labeling for Yap in the injured and noninjured areas of P4 utricular sensory epithelium after 24 hr in culture.
- https://doi.org/10.7554/eLife.25681.024
-
Figure 4—source data 3
The number cells doubly positive for EdU and Sox2 in the injured areas of P4 utricular sensory epithelium infected with virus carrying GFP or GFP-YTIP.
The numbers demonstrated for GFP-positive and GFP-negative cellular populations.
- https://doi.org/10.7554/eLife.25681.025
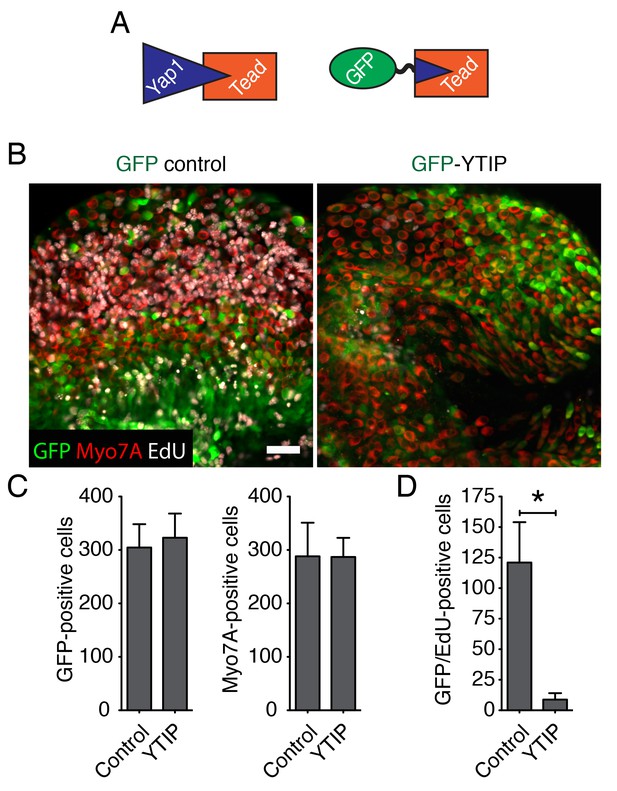
Yap controls supporting-cell proliferation in vitro.
(A) A schematic representation of Yap-Tead and GFP-YTIP-Tead interactions. Because Tead can no longer interact with the endogenous Yap protein in the presence of the dominant-negative YTIP peptide, overexpression of GFP-YTIP abrogates nuclear Yap functions. (B) As demonstrated by EdU incorporation (white), cellular proliferation can be observed in the utricular sensory epithelia of E16.5 utricles in GFP virus-infected but not in GFP-YTIP virus-infected cultures. The scale bar represents 50 μm. (C) Quantification of GFP-positive cells and Myo7A-positive hair cells in the utricles in panel A shows no statistical significance (p>0.05; N = 4 for each). (D) The number of cells doubly positive for EdU and GFP is significantly greater in GFP cultures than in GFP-YTIP preparations (p<0.001; N = 4 for each).
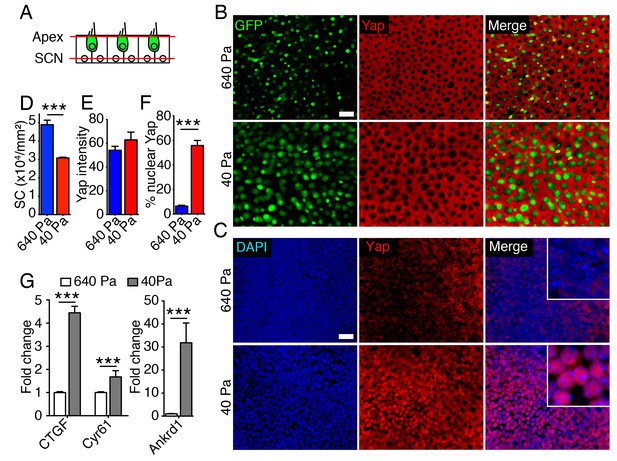
Subcellular localization of Yap in organotypic bubble cultures.
(A) In a schematic representation of a cross-section through an E17.5 Atoh-nGFP utricle, red lines represent optical sections through the apices of hair and supporting cells (Apex; shown in panel B) and through supporting cell nuclei (SCN; shown in panel C). Hair cells are marked in green. (B) In surface views of utricles maintained for 3 d as organotypic bubble cultures in collagen gels of Young's modulus 640 Pa (top panels) or 40 Pa (bottom panels), cytoplasmic Yap protein occurs in supporting cells but is nor observed in hair cells (green). The scale bar represents 25 μm. (C) In the same organotypic bubble cultures, Yap protein is excluded from the nuclei of supporting cells in a 640 Pa collagen gel (top panels). In contrast, Yap is translocated into the nuclei of supporting cells in a 40 Pa gel (bottom panels). The insets in the merge panels are magnified fourfold. The scale bar represents 25 μm. (D) Quantification of the density of Sox2-positive cells demonstrates a significant decrease in 40 Pa gels as compared to 640 Pa gels (means ± SEMs; p<0.001; N = 3 for each; Figure 5—source data 1). (E) The mean fluorescence intensity of antibody labeling for Yap is unchanged in utricles cultured for 3 d in 40 Pa gels as compared to 640 Pa gels (value range 0–85; means ± SEMs; p>0.05; N = 5 for 640 Pa and N = 6 for 40 Pa; Figure 5—source data 2). (F) The percentage of supporting cells showing Yap nuclear translocation is significantly higher in 40 Pa gels than in 640 Pa gels (means ± SEMs; p<0.0001; N = 4 for each; Figure 5—source data 3). (G) qPCR analysis of utricle bubble cultures maintained for 4 d reveals significant increase in the expression of the Yap target genes Ctgf, Cyr61, and Ankrd1 in 40 Pa gels as compared to 640 Pa gels (means ± SEMs; p<0.001 for each; N = 10 for each; Figure 5—source data 4).
-
Figure 5—source data 1
Quantification of the density of Sox2-positive cells in utricular bubble cultures maintained in 40 Pa and 640 Pa gels for 3 d.
- https://doi.org/10.7554/eLife.25681.028
-
Figure 5—source data 2
The mean fluorescence intensity of antibody labeling for Yap in the supporting-cell cytoplasm in utricular bubble cultures maintained in 40 Pa and 640 Pa gels for 3 d.
- https://doi.org/10.7554/eLife.25681.029
-
Figure 5—source data 3
The percentage of supporting cells showing Yap nuclear translocation in utricular bubble cultures maintained in 40 Pa and 640 Pa gels for 3 d.
- https://doi.org/10.7554/eLife.25681.030
-
Figure 5—source data 4
qPCR analysis of the expression of the Yap target genes Ctgf, Cyr61, and Ankrd1 in utricular bubble cultures maintained in 40 Pa and 640 Pa gels for 3 d.
- https://doi.org/10.7554/eLife.25681.031
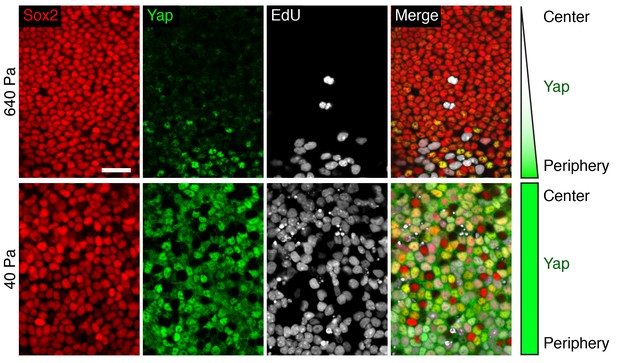
Subcellular Yap localization in organotypic bubble cultures.
In utricles maintained for 4 d as organotypic bubble cultures in 640 Pa collagen gels (top panels), nuclear Yap labeling (green) occurs in Sox2-positive supporting cells (red) at the periphery of the macula. EdU incorporation (white) demonstrates cell proliferation. In similar preparations of the utricles maintained in 40 Pa gels (lower panels), nuclear Yap labeling and EdU incorporation occur in the Sox2-positive supporting cells throughout the sensory epithelium. The scale bar represents 25 μm.
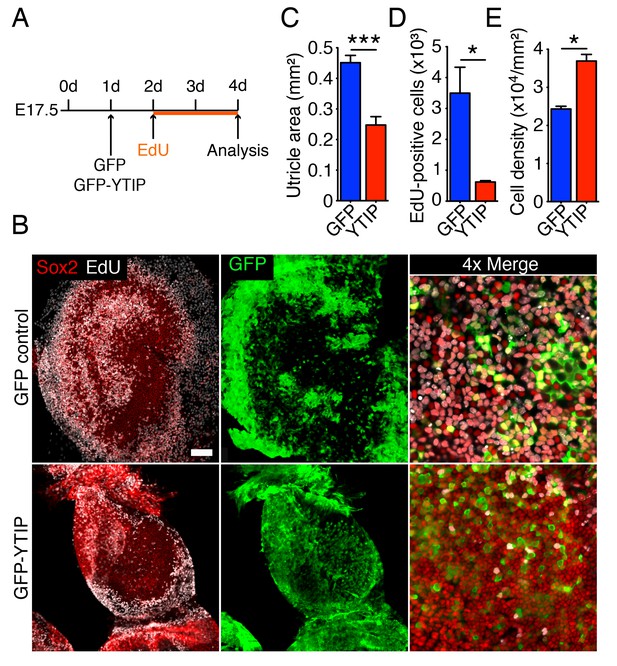
Yap controls utricular growth in organotypic bubble cultures.
(A) A schematic representation of an experiment on an organotypic bubble culture shows the time at which the utricles are transfected with virus expressing GFP or GFP-YTIP and the duration of EdU labeling. (B) Sox2 antiserum (red) labels the sensory epithelia of whole-mount preparations of utricular bubbles infected with virus expressing GFP or GFP-YTIP. As demonstrated by the incorporation of EdU (white), supporting cells in E17.5 utricles in GFP virus-infected cultures, but not in GFP-YTIP virus-infected cultures, re-enter the cell cycle. GFP, which demonstrates infected cells, is shown in green. The scale bar represents 100 μm. (C) The area of the sensory epithelia and (D) the number of EdU-positive cells in GFP virus-infected controls are increased significantly as compared to GFP-YTIP virus-infected utricles (means ± SEMs; area p<0.001; cell number p<0.05; N = 4 for GFP and N = 3 for GFP-YTIP; Figure 6—source data 1 and 2). (E) The extent of proliferation is inversely correlated with supporting-cell density, which is significantly decreased in GFP virus-infected controls as compared to GFP-YTIP virus-infected utricles (means ± SEMs; p<0.05; N = 5 for GFP and N = 6 for GFP-YTIP; Figure 6—source data 3).
-
Figure 6—source data 1
The area of the sensory epithelia in GFP and GFP-YTIP virus-infected utricular bubble cultures maintained in 40 Pa gels for 4 d.
- https://doi.org/10.7554/eLife.25681.034
-
Figure 6—source data 2
The number of EdU-positive supporting cells in GFP and GFP-YTIP virus-infected utricular bubble cultures maintained in 40 Pa gels for 4 d.
- https://doi.org/10.7554/eLife.25681.035
-
Figure 6—source data 3
The supporting-cell density in GFP and GFP-YTIP virus-infected utricular bubble cultures maintained in 40 Pa gels for 4 d.
- https://doi.org/10.7554/eLife.25681.036
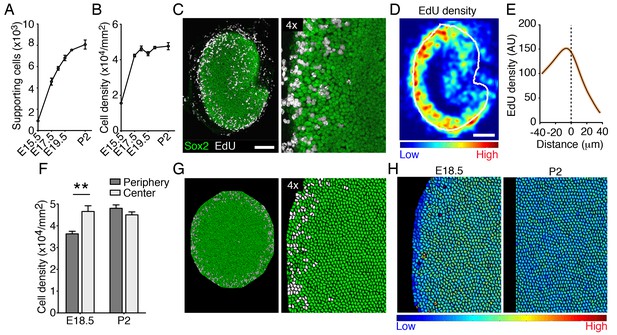
Cellular density and proliferation in the developing utricle.
(A) The numbers of supporting cells in utricular sensory epithelia increases significantly from E15.5 to P2 (means ± SEMs; p<0.05, N = 5 for each; Figure 7—source data 1). (B) The supporting-cell density grew significantly from E15.5 to E17.5 (means ± SEMs; p<0.05, N = 5 for each; Figure 7—source data 2) and plateaus thereafter. (C) In the Sox2-positive utricular sensory macula (green) of an E18.5 embryo exposed to EdU 12 hr earlier, dividing supporting cells (white) can be seen at the organ's periphery. The scale bar represents 100 μm. (D) The intensity of EdU labeling and the outline of the utricular macula (white line) are averaged for five E18.5 utricles to demonstrate the consistent pattern of supporting-cell proliferation at the corresponding developmental stage. (E) The distribution of EdU intensity shown in (D) demonstrates a clear peak that lies immediately inward from the macular boundary (dashed line). The standard errors are less that 1% and are shown by the orange area around the curve (Figure 7—source data 3). (F) The supporting-cell density at E18.5 is significantly higher at the center of the utricular macula than that at the organ's periphery (means ± SEMs; p<0.01, N = 4 for each; Figure 7—source data 4). There is no significant difference in supporting-cell density between the periphery and the center of the macula at P2 (means ± SEMs; p>0.05; N = 4 for each; Figure 7—source data 4). (G) Simulations of EdU labeling in the elasticity-limited model at E18.5 resemble the distribution of proliferating supporting cells seen experimentally at the same developmental stage. (H) The same simulation demonstrates the gradient in internal cell pressure in the E18.5 utricle. Areas of low pressure, and therefore of low cellular density, occur at the periphery; high pressure is predicted at the macular center. The gradient in cellular pressure disappears at P2.
-
Figure 7—source data 1
The numbers of supporting cells in utricular sensory epithelia counted at different developmental stages.
- https://doi.org/10.7554/eLife.25681.038
-
Figure 7—source data 2
The supporting-cell density in utricular sensory epithelia at different developmental stages.
- https://doi.org/10.7554/eLife.25681.039
-
Figure 7—source data 3
The average distribution of EdU intensity along the outline of the utricular macula in E18.5 utricle.
- https://doi.org/10.7554/eLife.25681.040
-
Figure 7—source data 4
The supporting-cell density at E18.5 and P2 at the center and the periphery of the utricular macula.
- https://doi.org/10.7554/eLife.25681.041
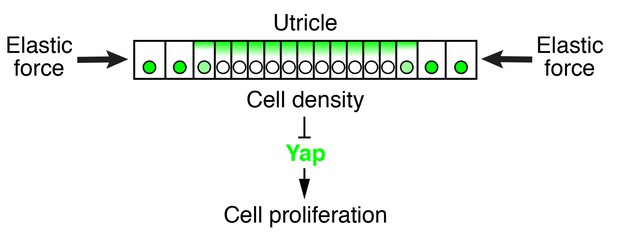
Elastic force restricts supporting-cell proliferation in the developing utricle.
A schematic drawing portrays the sensory epithelium expanding in an elastic matrix. The force opposing utricle growth creates a gradient of cellular density, with higher values at the center of the organ and lower ones in the periphery. A high density triggers the cytoplasmic localization and degradation of Yap, whose consequent depletion from nuclei results in cell-cycle arrest.
Videos
A simulation of the elasticity-limited model corresponding to Figure 1B spans developmental stages E15.5 to P14 at 1 hr intervals.
https://doi.org/10.7554/eLife.25681.010A simulation of the morphogen-limited model corresponding to Figure 1C spans developmental stages E15.5 to P14 at 1 hr intervals.
The concentration of the diffusible morphogen is shown on a spectral scale from blue as zero to red as the maximum, 0.20 pM. The white curve bounds the region in which a concentration exceeding the threshold value = 0.04 pM inhibits cellular proliferation.
A simulation of the theoretical model of macular development driven by a stem-cell population.
In this variant of the elasticity-limited model, a patch of stem cells (blue) proliferates and differentiates into supporting cells (green). The model assumes that only stem cells divide (white). The model predicts that stem cells at the center of the utricular macula can proliferate only briefly before cellular density increases and precludes further divisions. Although some stem cells are advected towards the macular periphery, the pattern of proliferation is not consistent with that observed in vivo.
Tables
Parameter values for simulations of cellular proliferation in the utricle.
| Description | Parameter name | Value |
|---|---|---|
| System size | 600 × 600 (hexagonal pixels) | |
| Amplitude of cell-membrane fluctuations | 5 (dimensionless) | |
| Cellular target volume | 40 pixels | |
| Cellular elastic modulus | 9 (dimensionless) | |
| Cellular target surface area | 25 pixels | |
| Cellular membrane elastic modulus | 1.5 (dimensionless) | |
| Elastic matrix’s Young’s modulus Elasticity-limited model Morphogen-limited model | 0.069 (dimensionless) 0.055 (dimensionless) | |
| Cell-cell interaction energy | 20 (dimensionless) | |
| Morphogen diffusion coefficient | 1.19·10−14 m2·s−1 | |
| Morphogen secretion rate | 0.23 s−1 | |
| Morphogen concentration threshold | 0.1 pM | |
| Time after division for cells to become quiescent | 2000 (Monte Carlo steps) | |
| Mean refractory time | 200 (Monte Carlo steps) | |
| Refractory time standard deviation | 50 (Monte Carlo steps) | |
| Monte Carlo step time | 210 s | |
| Model’s spatial scale | 8.6·10−13 m2 |
Expression ratios for relevant genes in utricular sensory epithelia in late embryonic development (E17.5) and after maturation (P9).
| Gene | RPKM E17.5 | RPKM P9 | Expression ratio E17.5/P9 | p-value | Function |
|---|---|---|---|---|---|
| Dach1 | 12.0 | 5.7 | 2.0 | <0.05 | Cytoskeletal interactors |
| Nf2 | 25.0 | 27.4 | 0.91 | >0.05 | |
| Wwc1 | 76.2 | 72.5 | 1.0 | >0.05 | |
| Sav1 | 17.8 | 27.2 | 0.65 | >0.05 | Kinases |
| Stk3 | 18.3 | 19.7 | 0.92 | >0.05 | |
| Stk4 | 13.6 | 11.4 | 1.1 | >0.05 | |
| Lats1 | 26.5 | 26.5 | 1.0 | >0.05 | |
| Lats2 | 5.2 | 3.6 | 1.4 | >0.05 | |
| Mob1a | 27.1 | 20.9 | 1.2 | >0.05 | |
| Mob1b | 13.9 | 17.5 | 0.79 | >0.05 | |
| Taz | 21.5 | 29.8 | 0.72 | >0.05 | Transcriptional coactivators |
| Yap1 | 53.9 | 63.5 | 0.84 | >0.05 |
-
The complete data set is available through GEO database accession number GSE72293. RPKM, reads per kilobase of transcript per million mapped reads.
Additional files
-
Source code 1
CompuCell 3D (Swat et al., 2012) scripts and parameter files for the elasticity-limited model.
- https://doi.org/10.7554/eLife.25681.044
-
Source code 2
CompuCell 3D (Swat et al., 2012) scripts and parameter files for the morphogen-limited model.
- https://doi.org/10.7554/eLife.25681.045
-
Source code 3
CompuCell 3D (Swat et al., 2012) scripts and parameter files for the stem cell model.
- https://doi.org/10.7554/eLife.25681.046
-
Source code 4
LabVIEW scripts to control the MP-285 micromanipulator for measuring the stiffness of collagen gels.
- https://doi.org/10.7554/eLife.25681.047





