A large gene family in fission yeast encodes spore killers that subvert Mendel’s law
Figures
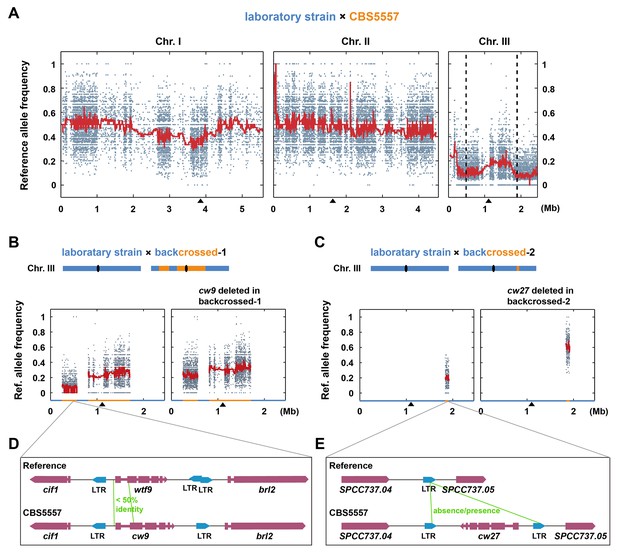
The identification of cw9 and cw27 genes as spore killers.
(A) NGS-assisted bulk segregant analysis of viable progenies from a cross between DY9974, a heterothallic derivative of CBS5557, and DY8531, a laboratory strain harboring the 2.23 Mb chromosome I inversion (Hu et al., 2015). Reference allele frequencies at SNP positions are plotted as dots. Red trend lines are based on a rolling median calculation. Two vertical dashed lines denote the positions of cw9 and cw27 genes on chromosome III. Black triangles mark the positions of centromeres. (B and C) Bulk segregant analysis of viable progenies from crosses between a laboratory strain and the backcrossed-1 strain, and between a laboratory strain and the backcrossed-2 strain, respectively. Blue and orange colored segments in the diagrams and on the X-axes of the plots denote chromosome III regions with reference genome sequence and those with CBS5557 genome sequence, respectively, in the two backcrossed strains. (D and E) Schematics of the regions surrounding cw9 and cw27 in the CBS5557 genome and the corresponding regions in the reference genome. Gene structures of cw9 and cw27 were predicted using the AUGUSTUS web server (Stanke et al., 2008). Solo LTRs were annotated based on BLAST analysis.
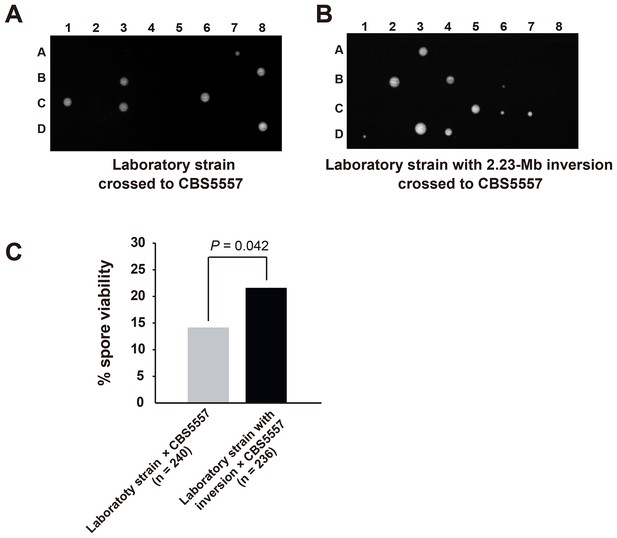
Hybrid sterility between CBS5557 and the laboratory strain can be partially rescued by eliminating the difference in the 2.23 Mb chromosome I inversion.
(A) Representative tetrads from a cross between LD775, an h- laboratory strain, and DY9974, an h+ derivative of CBS5557. (B) Representative tetrads from a cross between DY8531, an h- laboratory strain harboring the 2.23 Mb chromosome I inversion (Hu et al., 2015), and DY9974. (C) Quantitation of spore viability assessed by tetrad analysis. p-value was calculated using Fisher’s exact test. Numerical data are provided in Supplementary file 1.
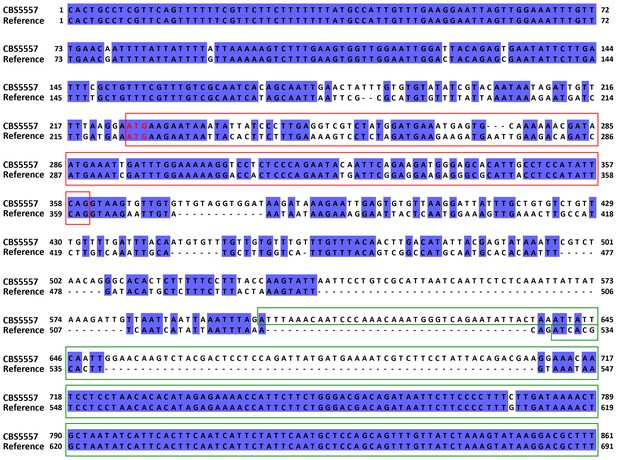
Alignment of the 5’ portions of the cw9 gene of CBS5557 and the wtf9 gene of the reference genome.
The sequences were aligned using MAFFT via Jalview (Katoh et al., 2002; Waterhouse et al., 2009). Identical bases are indicated by blue background. Red and green boxes denote the first and second predicted protein coding exons, respectively. Predicted start codons are highlighted with red letters. Gene structure of cw9 was predicted using the AUGUSTUS web server (Stanke et al., 2008).

Alignment of the two directly oriented LTRs flanking cw27 in the CBS5557 genome (top and bottom sequences, respectively) and the LTR at the corresponding location in the reference genome (middle sequence).
The reference genome LTR was referred to as PCRW_055 or C3_1895301 in Bowen et al. (2003). According to the classification proposed in Bowen et al., these three LTR sequences belong to the ζ clade. The sequences were aligned using MAFFT via Jalview. Identical bases are indicated by grey background. 7-nt-long homology at the breakpoint is highlighted in red. Matched sequences upstream and downstream of the breakpoint are colored in green.
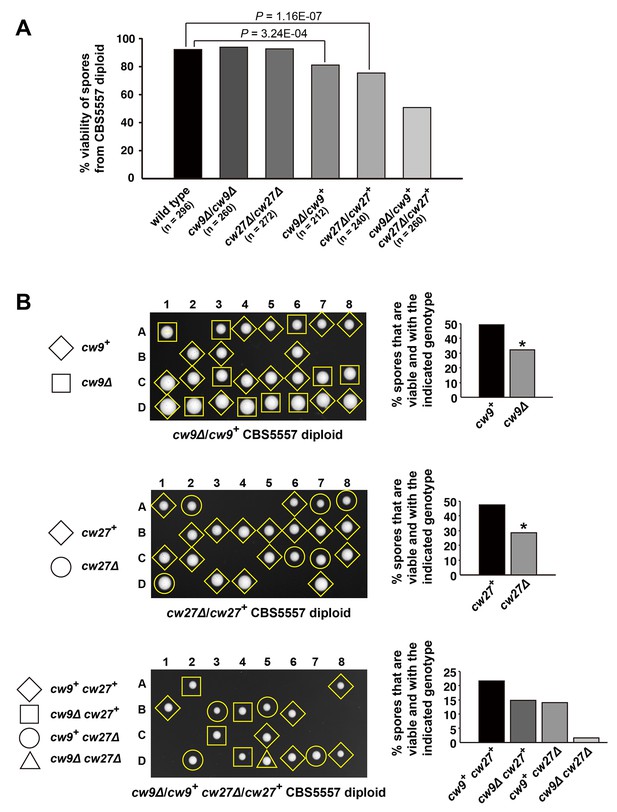
cw9 and cw27 act as spore killers when in the heterozygous state in the CBS5557 background.
(A) Heterozygous deletion but not homozygous deletion of cw9 or cw27 in CBS5557 h+/h- diploid caused spore viability loss. Spore viability was measured using tetrad analysis. Representative tetrads are shown in Figure 2—figure supplement 1 and in panel B. p-values were calculated using Fisher’s exact test. Numerical data are provided in Supplementary file 1. (B) Among the spores derived from CBS5557 diploids with heterozygous deletion, loss of viability mainly occurred to spores with deletion. Asterisks indicate significant deviation from 50% (p=1.94E-7 and 1.42E-11 for cw9∆ spores from cw9∆/cw9+ diploid and cw27∆ spores from cw27∆/cw27+ diploid, respectively, exact binomial test). Numerical data are provided in Supplementary file 1.
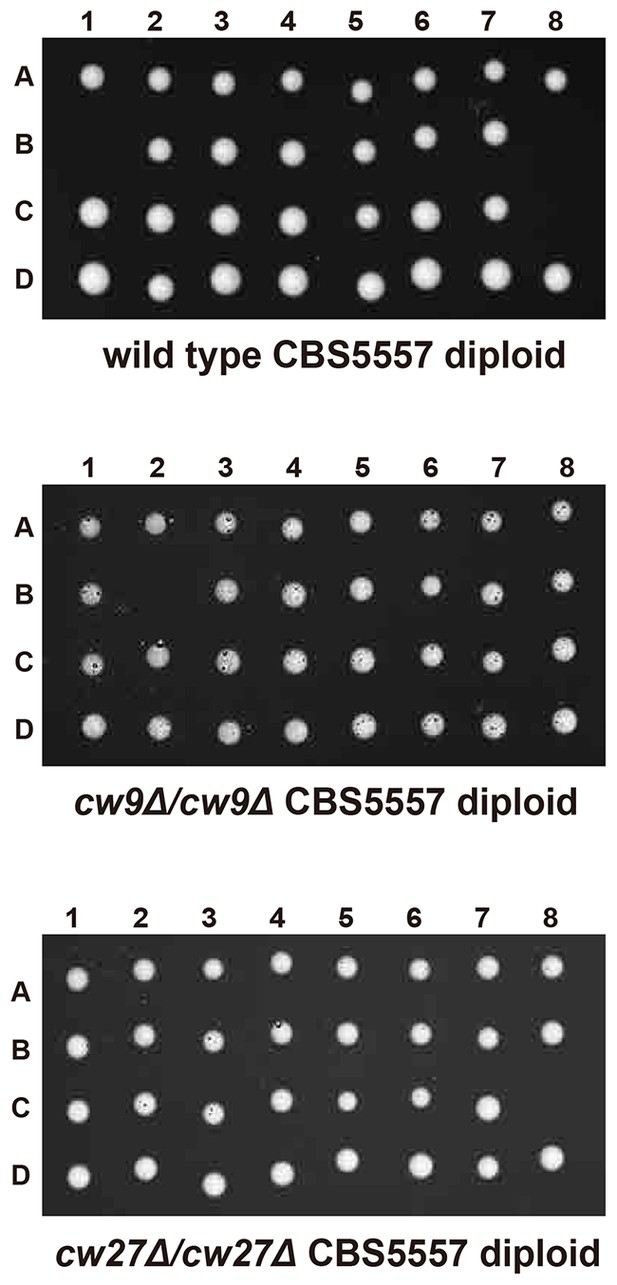
Representative tetrads from a wild type CBS5557 h+/h- diploid strain (DY21782), a CBS5557 h+/h- diploid strain with homozygous cw9 deletion (DY21838), and a CBS5557 h+/h- diploid strain with homozygous cw27 deletion (DY21842).
https://doi.org/10.7554/eLife.26057.008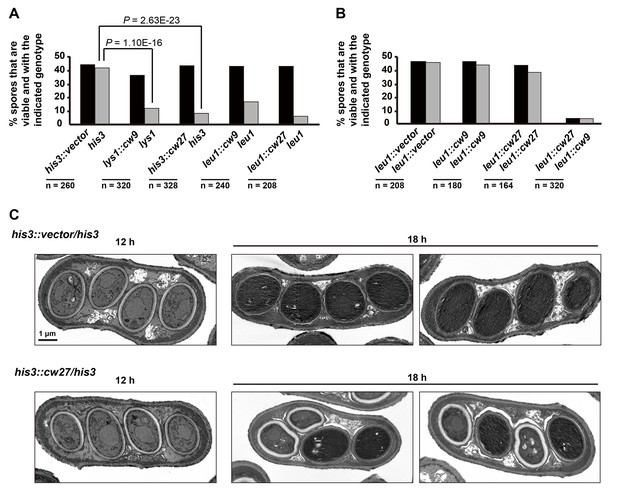
cw9 and cw27 act as spore killers when inserted into the genome of the laboratory strain.
(A) In crosses of laboratory strains, when only one parental strain had an insertion of cw9 or cw27 at his3, lys1, or leu1 locus, spores without the insertion suffered viability loss. Representative tetrads are shown in Figure 3—figure supplement 1. p-values were calculated using Fisher’s exact test. Numerical data are provided in Supplementary file 1. (B) In crosses of laboratory strains, when both parental strains had an insertion of cw9 or cw27 at the leu1 locus, spore viability was normal when parents had the same killer, but was severely low when parents had different killers. The two parental alleles were distinguished by leu1-linked antibiotic resistance markers. Representative tetrads are shown in Figure 3—figure supplement 2. Numerical data are provided in Supplementary file 1. (C) Electron microscopy analysis of laboratory-background h+/h- diploid cells undergoing synchronous meiosis and sporulation.
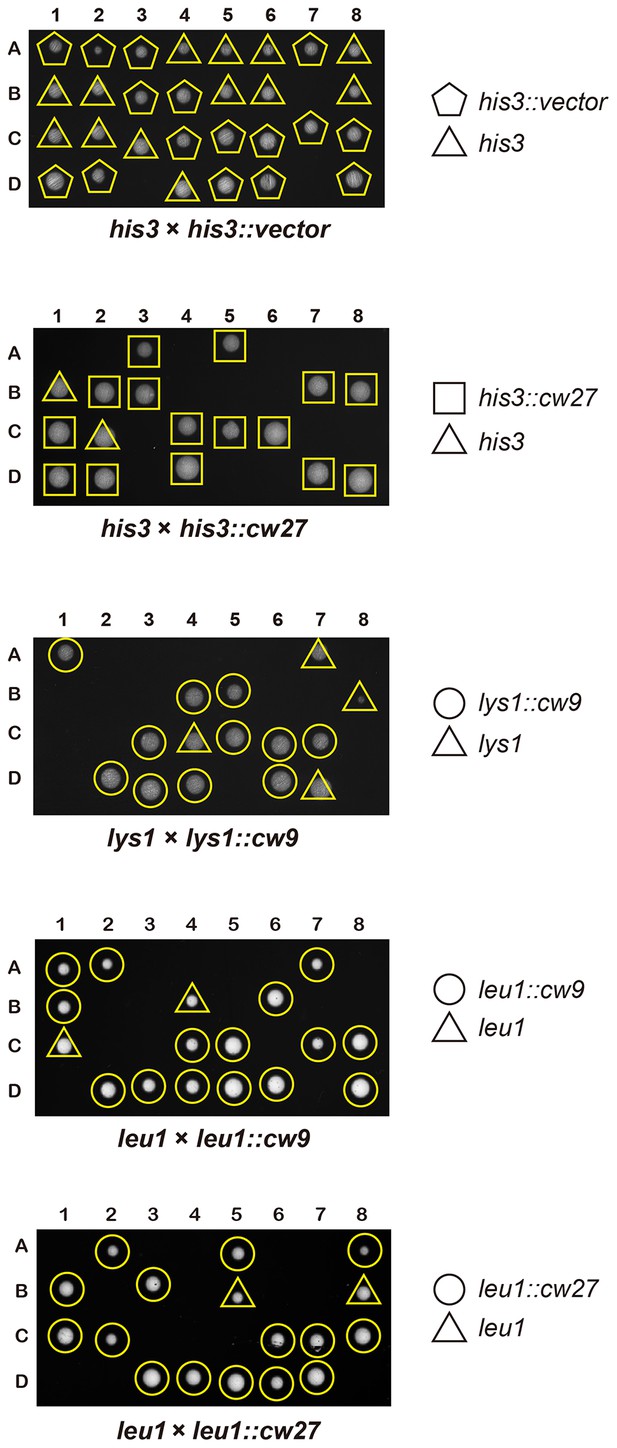
Representative tetrads from laboratory-background crosses in which only one of the parental haploid strains had a vector or a killer-containing plasmid integrated at the his3, lys1, or leu1 locus.
https://doi.org/10.7554/eLife.26057.010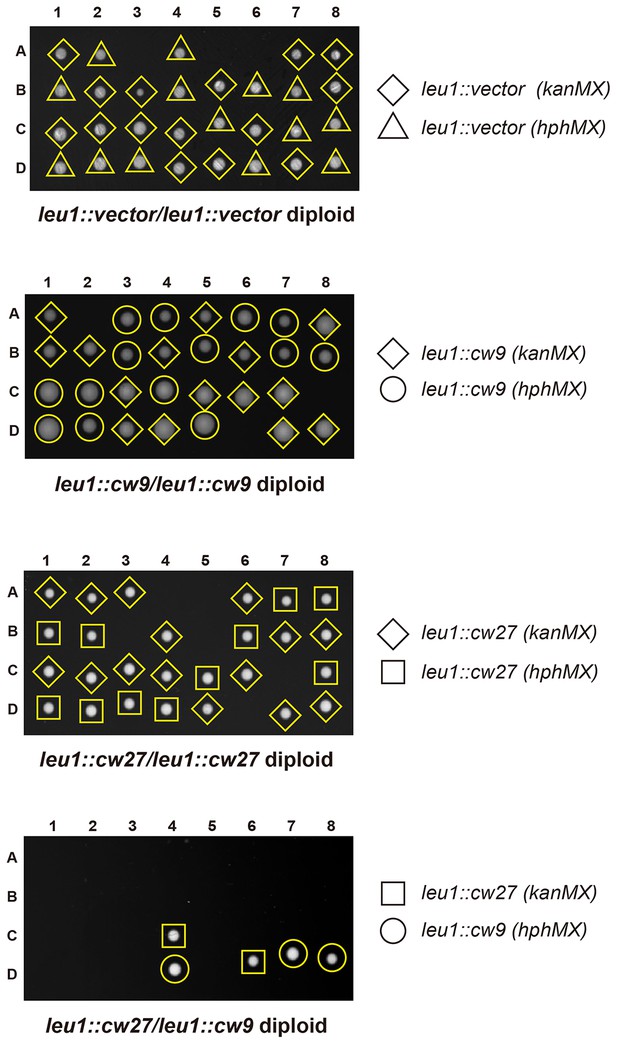
Representative tetrads from three laboratory-background h+/h- diploid strains homozygous for plasmid integration at the leu1 locus and a laboratory-background h+/h- diploid strain heterozygous for plasmid integration at the leu1 locus, with one allele containing cw9 and the other allele containing cw27.
kanMX and hphMX markers were inserted between the coordinates 1963090 and 1963096 on chromosome II so that they are tightly linked to leu1.
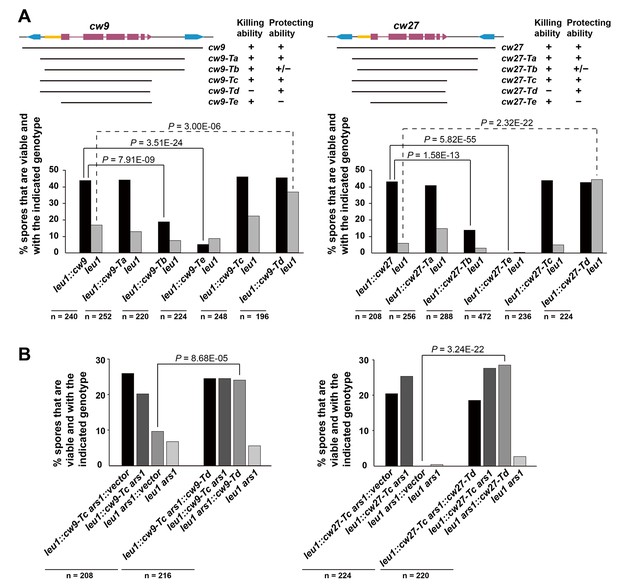
Sequence requirement for the killing and the protecting activities of cw9 and cw27.
(A) Truncation analysis to assess the involvement of the 5’ and 3’ sequences of cw9 and cw27 in spore killing. In the diagrams on top, blue arrows represent LTRs and yellow bars represent the conserved_up sequence. Representative tetrads are shown in Figure 4—figure supplement 1. p-values were calculated using Fisher’s exact test. Numerical data are provided in Supplementary file 1. (B) The Td versions of cw9 and cw27 are able to effectively protect against killing despite their lack of killing activity. Representative tetrads are shown in Figure 4—figure supplement 2. p-values were calculated using Fisher’s exact test. Numerical data are provided in Supplementary file 1.
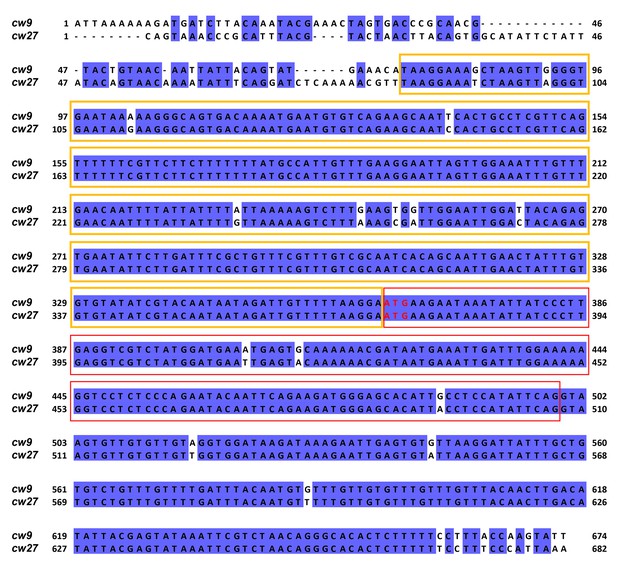
Alignment of the 5’ portions of cw9 and cw27.
The starting positions of these two sequences correspond to those of the Ta truncations. The sequences were aligned using MAFFT via Jalview. Identical bases are indicated by blue background. Yellow and red boxes denote the conserved_up sequence and the first predicted protein coding exon, respectively. Predicted start codons are highlighted with red letters.
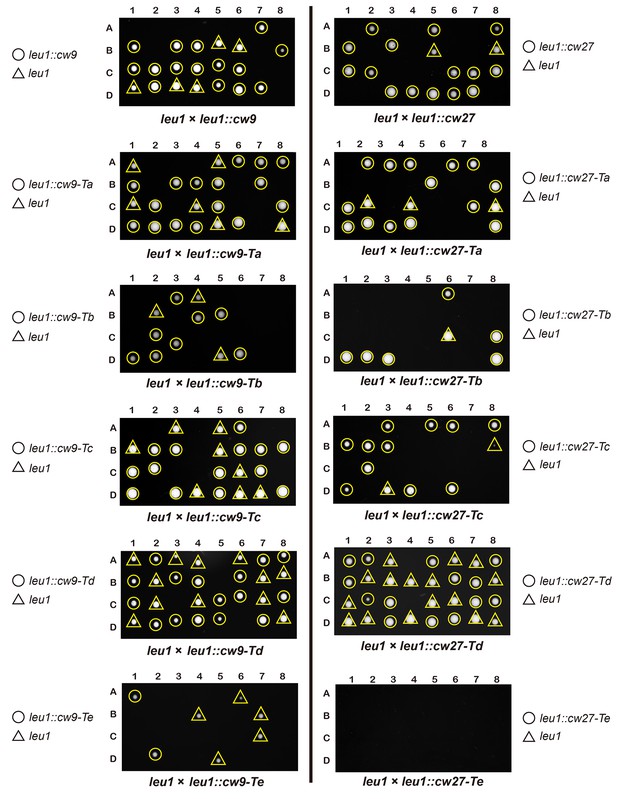
Representative tetrads from laboratory-background h+/h- diploid strains heterozygous for plasmid integration at the leu1 locus.
https://doi.org/10.7554/eLife.26057.014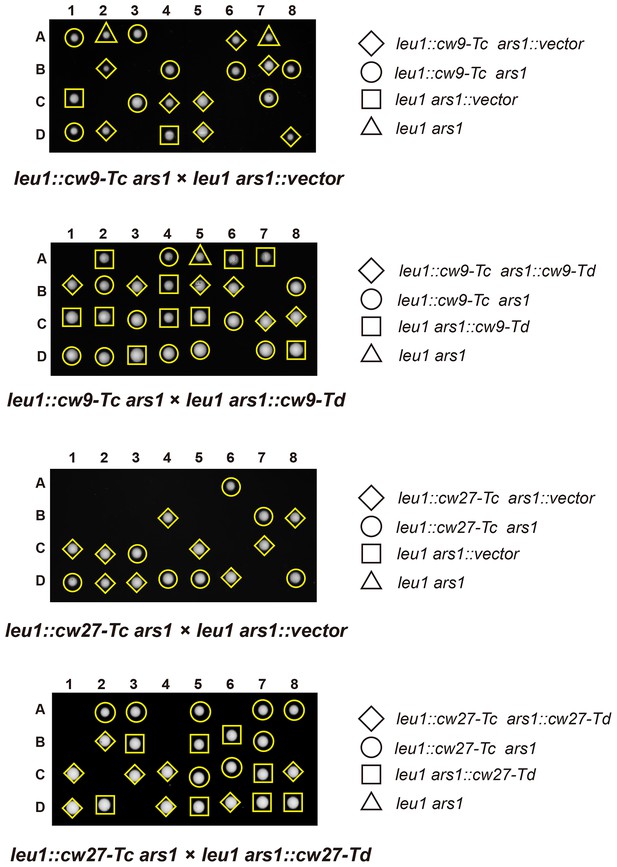
Representative tetrads from laboratory-background h+/h- diploid strains heterozygous for a Tc-version killer-containing plasmid integrated at the leu1 locus on chromosome II, and also heterozygous for a vector or a Td-version killer-containing plasmid integrated at the ars1 replication origin region upstream of the hus5 gene on chromosome I.
https://doi.org/10.7554/eLife.26057.015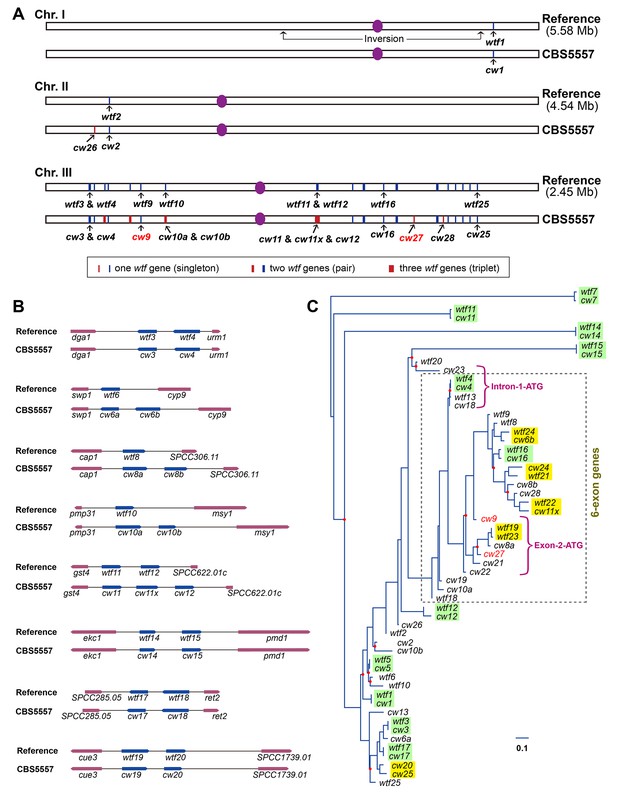
wtf genes vary both in numbers and sequences between the reference genome and the CBS5557 genome.
(A) Genomic locations of wtf genes in the reference genome and in the CBS5557 genome. The 25 wtf genes in the reference genome have been named according to their order in the genome (Bowen et al., 2003). Their locations are depicted as 20 blue vertical bars, including five thick bars denoting five tandem pairs. Our PacBio sequencing analysis revealed that in the CBS5557 genome, there are 32 wtf genes, whose locations are depicted as 23 vertical bars, including three thin red bars at locations where no wtf genes exist in the reference genome and four thick red bars at locations where compared to the reference genome one extra wtf gene is found. The wtf genes in the CBS5557 genome are named with the prefix cw and a number from the name of the syntenic gene in the reference genome. Two genes of a tandem pair corresponding to a singleton in the reference genome are distinguished using the suffixes a and b. Among the three genes of the triplet, two are named cw11 and cw12 based on their homology to wtf11 and wtf12, respectively, and the gene situated between cw11 and cw12 is named cw11x. Genes at new locations are named cw26, cw27, and cw28. Chromosome lengths are not drawn to scale. (B) Diagrams depicting the eight genomic locations with more than one wtf gene in at least one of the two genomes. Genes are shown as arrows. Introns are not shown. wtf17 is depicted not according to its annotation at PomBase, with its 5’ boundary revised based on sequence alignment. (C) Maximum likelihood phylogenetic tree of 57 wtf genes of the reference genome and the CBS5557 genome. DNA sequences including the conserved_up regions, predicted coding sequences, and associated introns were aligned using the L-INS-i iterative refinement algorithm of MAFFT (Katoh and Standley, 2014) (Figure 5—source data 1). Maximum likelihood analysis was performed using IQ-TREE (Nguyen et al., 2015). The tree was rooted by midpoint rooting (Hess and De Moraes Russo, 2007). Red dots on nodes indicate IQ-tree-calculated ultrafast bootstrap (UFBoot) support values < 95%. Colored rectangles highlight phylogenetic neighbors. Two genes are considered phylogenetic neighbors if they are separated by a single internal node with a support value >= 95%. Green rectangles indicate the 11 pairs of neighbors each composed of a reference wtf gene and a syntenic CBS5557 wtf gene. Yellow rectangles indicate the five pairs each composed of two wtf genes locating at different genomic positions. Magenta brackets denote the 12 genes that share a 150 bp sequence within the predicted intron 1 (see Figure 5—figure supplement 3). These 12 genes are divided into two subtypes, Intron-1-ATG genes and Exon-2-ATG genes, based on the locations of the first ATG codons downstream of the 150-bp-long sequence. The brown dashed box indicates genes with six exons. Scale bar, 0.1 nucleotide substitutions per nucleotide site.
-
Figure 5—source data 1
MAFFT-aligned DNA sequences of 57 wtf genes of the reference genome and the CBS5557 genome.
The sequences include the conserved_up regions, predicted coding sequences, and associated introns.
- https://doi.org/10.7554/eLife.26057.017
-
Figure 5—source data 2
Gene structure predictions for 57 wtf genes of the reference genome and the CBS5557 genome.
- https://doi.org/10.7554/eLife.26057.018
-
Figure 5—source data 3
MAFFT-aligned amino acid sequences of the predicted protein products of 57 wtf genes of the reference genome and the CBS5557 genome.
- https://doi.org/10.7554/eLife.26057.019
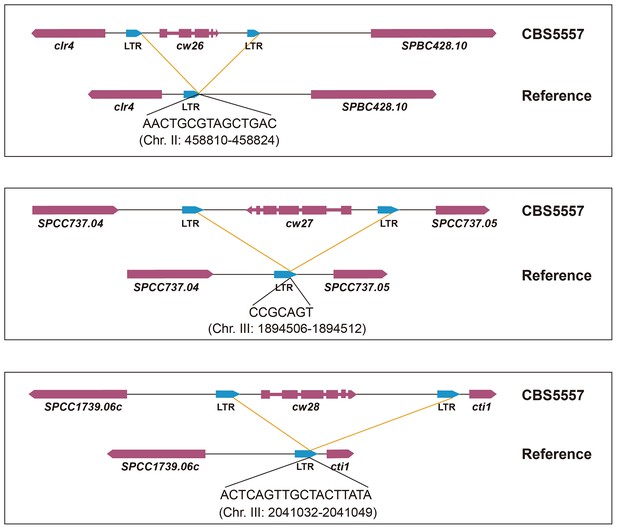
Schematics of how the three CBS5557-only singleton wtf genes may have been lost in the laboratory strain through LTR-mediated recombination.
Shown at the bottom of each diagram is the breakpoint junction sequence with 100% identity between the two directly oriented LTRs flanking the CBS5557-only singleton wtf gene and its position in the reference genome.
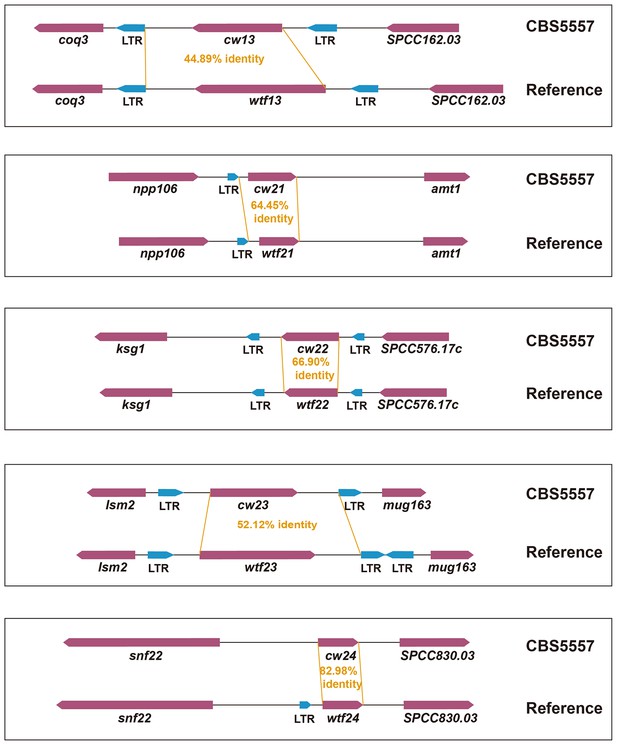
Schematics depicting the high levels of diversity between five singleton CBS5557 wtf genes and their counterparts in the reference genome.
Genes are shown as maroon arrows. Introns are not shown. Sequence alignment was performed using MAFFT via Jalview. Pair-wise identity was calculated by dividing the numbers of identical bases by the alignment length.
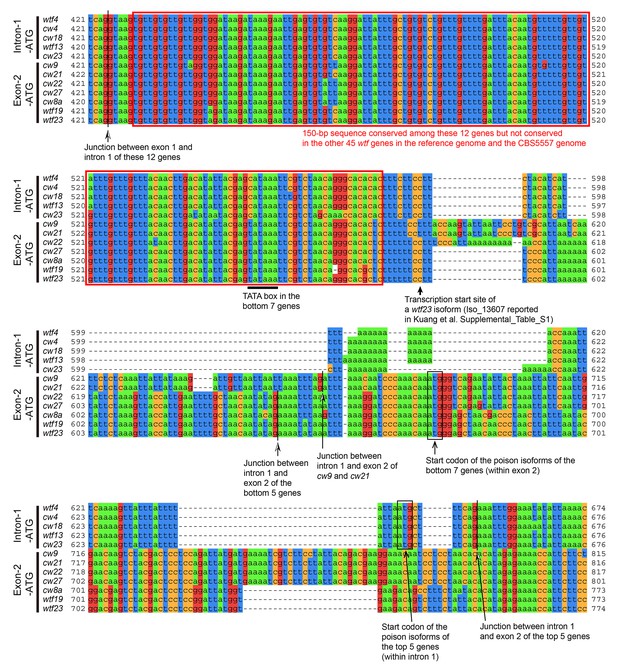
cw9, cw27, and 10 other wtf genes in the reference genome and the CBS5557 genome share a 150 bp conserved sequence in the predicted intron 1.
Red boxes denote the 150 bp sequence. The transcription start site of a short wtf23 isoform identified by the Iso-Seq method (Kuang et al., 2017) is situated closely downstream of this 150 bp sequence, and at an optimal distance from a canonical TATA box (TATAAA) (Li et al., 2015). The first ATG codons downstream of the 150 bp sequence are located within the predicted intron 1 of the top five genes, which we call Intron-1-ATG genes, and within the predicted exon 2 of the bottom 7 genes, which we call Exon-2-ATG genes. These ATG codons are all in frame with the predicted coding sequences. Proteins initiated from these ATG codons correspond to the poison isoforms in the model proposed by Nuckolls et al., 2017). Sequence coordinates and alignment are based on the MAFFT-aligned DNA sequences in Figure 5—source data 1.
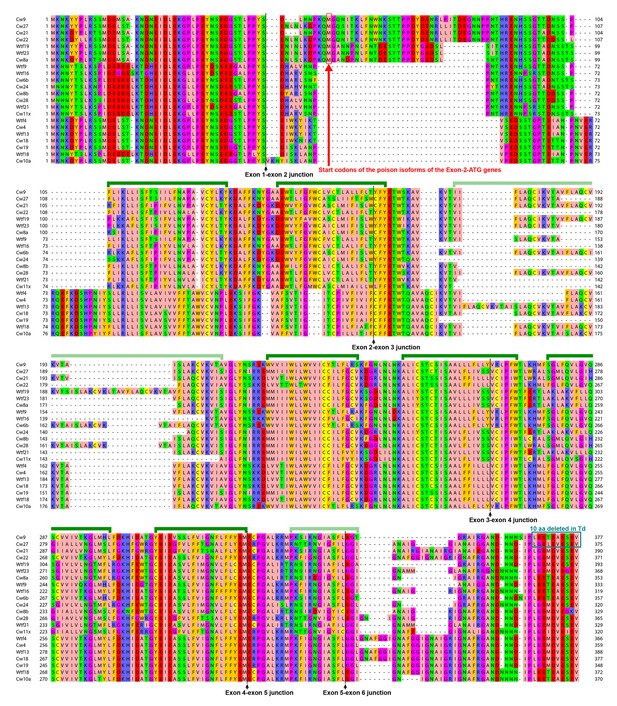
The predicted protein products of 6-exon-containing wtf genes in the reference genome and the CBS5557 genome.
Alignment is based on the MAFFT-aligned protein sequences in Figure 5—source data 3. Protein products of wtf8, wtf22, wtf24, and cw16, which belong to the 6-exon group but appear to have suffered pseudogenizing mutations, are not shown. Green brackets denote the eight transmembrane helices predicted by PolyPhobius (http://phobius.sbc.su.se/poly.html) (Käll et al., 2005). Helices 3 and 8 are indicated with light green brackets, because the former is absent in some of the aligned sequences, and the latter has low residue-wise posterior probabilities in the PolyPhobius prediction.
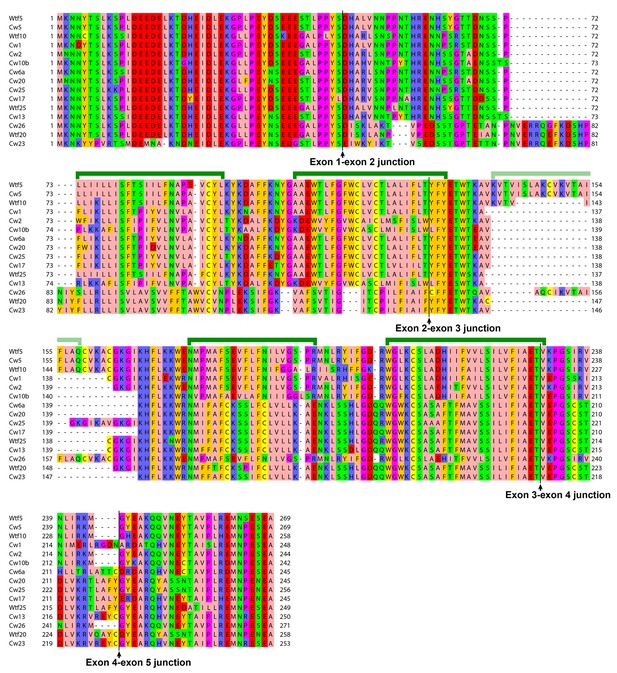
The predicted protein products of 5-exon-containing wtf genes in the reference genome and the CBS5557 genome.
Alignment is based on the MAFFT-aligned protein sequences in Figure 5—source data 3. Protein products of the eight most divergent genes (wtf7, wtf11, wtf14, wtf15, cw7, cw11, cw14, and cw15) and eight genes that appear to have suffered pseudogenizing mutations (wtf1, wtf2, wtf3, wtf6, wtf12, wtf17, cw3, and cw12) are not shown. Green brackets denote the five transmembrane helices predicted by PolyPhobius. Helix three is indicated with a light green bracket, because it is absent in some of the aligned sequences.
Additional files
-
Supplementary file 1
Numerical data of the tetrad analysis.
- https://doi.org/10.7554/eLife.26057.025
-
Supplementary file 2
Fission yeast strains used in this study.
- https://doi.org/10.7554/eLife.26057.026





