Synaptic and peptidergic connectome of a neurosecretory center in the annelid brain
Figures
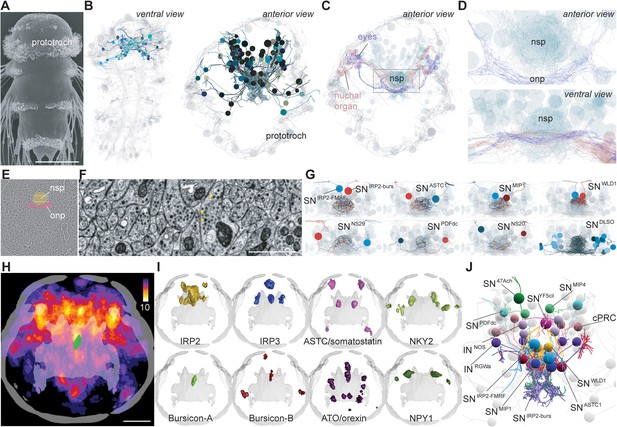
EM reconstruction and mapping of proneuropeptide expression in the apical nervous system of Platynereis larvae.
(A) Scanning electron micrograph of a 3-day-old Platynereis larva. (B) Reconstructed apical nervous system neurons (ANS) (shades of blue) in a full-body serial section transmission electron microscopy (ssTEM) dataset of a 3-day-old larva, shown against a framework of reconstructed ciliated cells and axonal scaffold (light grey). Axons and dendrites appear as lines and cell body positions are represented by spheres. (C) ANS neurons project to an apical neurosecretory plexus in the center of the head, which forms a small sphere dorsal and apical to the optic neuropil and nuchal organ neuropil. (D) Close-up view of neurosecretory plexus indicated by box in C. (E) TEM image of a section in the larval head. The neurosecretory plexus and the optic neuropil are highlighted. (F) Close-up view of the neurosecretory plexus indicated by boxed area in E. Yellow arrowheads point at dense core vesicles. (G) Examples of bilaterally symmetric pairs of sensory neurons that innervate different regions of the neurosecretory plexus in the reconstructed ANS (grey), ventral view. (H) Heat map of the expression of 58 proneuropeptides in the 3-day-old Platynereis larva, apical view, projected on a reference scaffold of cilia and axonal scaffold (grey). Color scheme indicates number of different proneuropeptides expressed in a cell. An apical bursicon-A-expressing neuron of the ANS (SNIRP2-burs) is overlaid for spatial reference (green). (I) Examples of average gene expression patterns of individual proneuropeptides from the 3 day proneuropeptide expression atlas. (J) ssTEM reconstruction of the 3-day-old larval ANS showing neurons that could be assigned specific proneuropeptide expression based on position and sensory cilia morphology. Abbreviations: nsp, apical neurosecretory plexus; onp, optic neuropil. Scale bars: A, 70 µm; H, 30 µm; F, 1 µm.
-
Figure 1—source data 1
Reconstructed skeletons of Platynereis neurons in the 3-day-old larva ssTEM data stack have been submitted to the NeuroMorpho database http://neuromorpho.org/dableFiles/Jekely/Supplementary/Williams_Archive.zip.
- https://doi.org/10.7554/eLife.26349.012
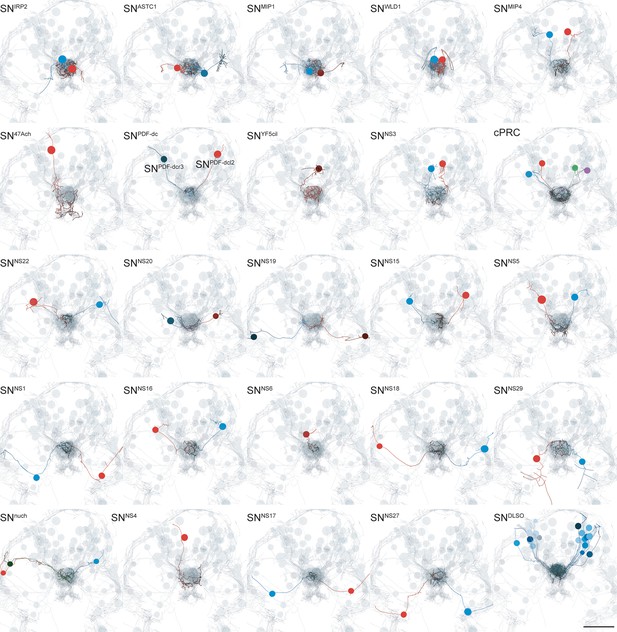
TEM reconstruction of ANS neurons.
Bilaterally symmetric pairs and single asymmetric sensory neurons are shown in anterior view. Axons and dendrites appear as lines and cell body positions are represented by spheres. The reconstructed ciliary band cells and all ANS cells (grey) are included to show the perimeter of the head. Scale bar: 30 µm.

Average gene expression patterns of individual proneuropeptides from a whole-body gene expression atlas from 2-day-old larvae, anterior view.
Scale bar: 30 µm.
-
Figure 1—figure supplement 2—source data 1
Blender gene expression atlas for 2-day-old Platynereis larvae, including gene expression patterns of 53 neuropeptide precursor genes.
Files with extension .blend to be opened in Blender (https://www.blender.org/); plus acetylated tubulin average scaffold and DAPI registration reference files (.tiff files).
- https://doi.org/10.7554/eLife.26349.005

Average gene expression patterns of individual proneuropeptides from a whole-body gene expression atlas from 3-day-old larvae, anterior view.
Scale bar: 30 µm.
-
Figure 1—figure supplement 3—source data 1
Blender gene expression atlas for 3-day-old Platynereis larvae, including gene expression patterns of 58 neuropeptide precursor genes.
Files with extension .blend to be opened in Blender (https://www.blender.org/); plus acetylated tubulin average scaffold and DAPI registration reference files (.tiff files).
- https://doi.org/10.7554/eLife.26349.007
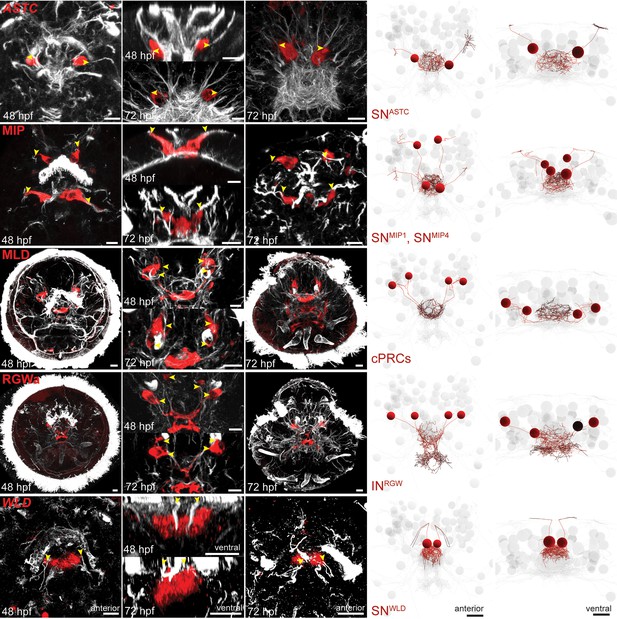
Stainings to assign molecular identity to ANS neurons based on their position, morphology and unique sensory cilia.
Gene expression patterns for allatostatin C (ASTC), myoinhibitory peptide (MIP), MLD peptide (pedal peptide 2), RGWa peptide and WLD peptide were established in 48 and 72 hpf larvae by immunostaining with a polyclonal antibody (MIP, MLD, RGWa), or in situ hybridization with an RNA probe (ASTC, WLD). All samples were counterstained with an antibody raised against acetylated tubulin to label cilia and axonal scaffold (white). Close-ups of stainings in anterior and ventral orientation are shown (panels with black background), for comparison with reconstructed neurons from the 72 hpf TEM dataset (panels with white background). Yellow arrowheads indicate the point at which sensory dendrites (white) emerge from the cell body (red) for ASTC-, MIP-, and WLD-expressing cells. For RGWa- and MLD-expressing cells, yellow arrowheads indicate cell bodies. Scale bars: 10 µm.
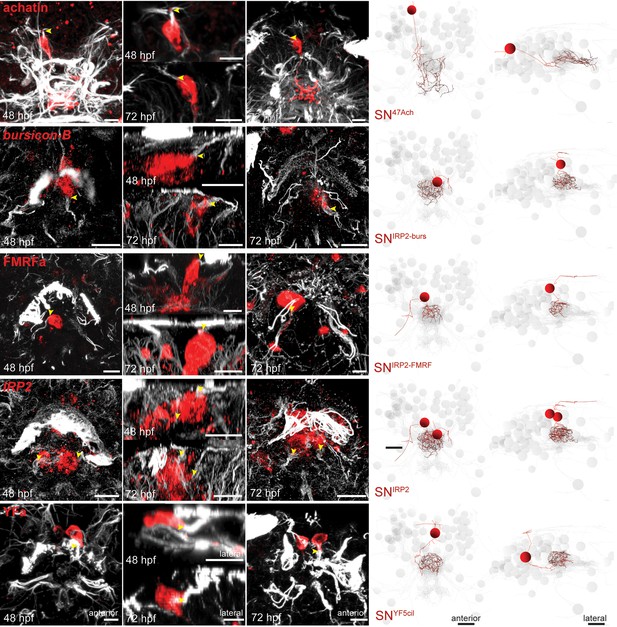
Stainings to assign molecular identity to ANS neurons based on their position, morphology and unique sensory cilia.
Gene expression patterns for achatin, bursicon-B, FMRFa peptide (FMRFa), insulin-related peptide (IRP2), and YFa peptide were established in 48 and 72 hpf larvae by immunostaining with a polyclonal antibody (achatin, FMRFa, YFa), or in situ hybridization with an RNA probe (bursicon-B, IRP2). All samples were counterstained with an antibody raised against acetylated tubulin to label cilia and axonal scaffold (white). Close-ups of stainings in anterior and right lateral orientation are shown (panels with black background), for comparison with reconstructed neurons from the 72 hpf TEM dataset (panels with white background). Yellow arrowheads indicate the point at which sensory dendrites (white) emerge from the cell body (red) for all cells. Scale bars: 10 µm.
-
Figure 1—figure supplement 5—source data 1
Videos of close-up immunostainings or in situ hybridizations counterstained with acetylated tubulin antibody for neuropeptides or neuropeptide precursors expressed in the central sensory neurons SNIRP2-bursand SNIRP2-FMRF.
The same sensory neurons have also been reconstructed by ssTEM, see Video 2.
- https://doi.org/10.7554/eLife.26349.010
-
Figure 1—figure supplement 5—source data 2
Videos of close-up immunostainings or in situ hybridizations counterstained with acetylated tubulin antibody for neuropeptides or neuropeptide precursors expressed in central sensory neurons SNMIP1, SNWLD and SNYF5cil.
The same sensory neurons have also been reconstructed by ssTEM, see Video 2.
- https://doi.org/10.7554/eLife.26349.011
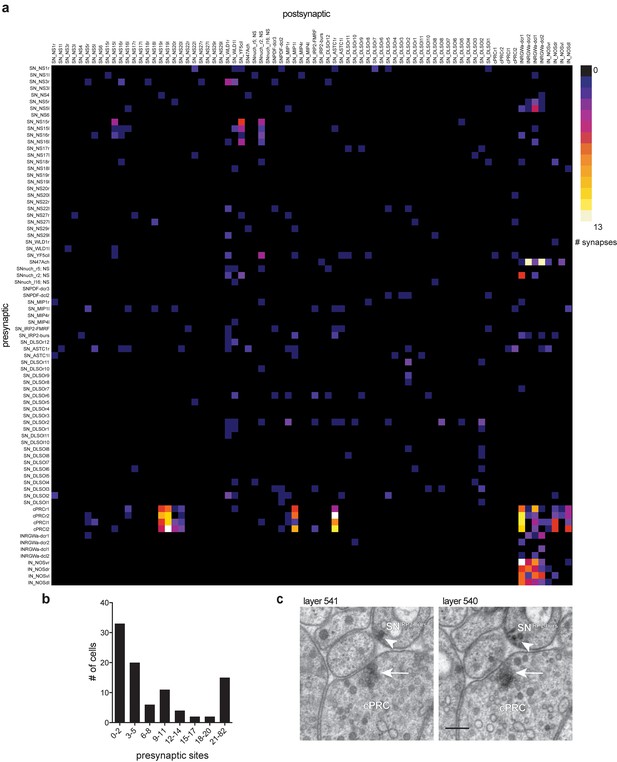
Low level of synaptic connectivity of ANS neurons.
(A) Full matrix of synaptic connectivity of ANS neurons. With the exception of the ciliary photoreceptors (cPRC), a few sensory cells, and the INRGW and INNOSprojection interneurons, the ANS shows very sparse synaptic connectivity. (B) Histogram of the number of presynaptic sites in ANS neurons. (C) Comparison of a cholinergic synapse of a ciliary photoreceptor (arrow) to a peptidergic synapse of the SNIRP2-burs cell (arrowhead). Two consecutive sections are shown. Scale bar in C, 200 nm. Spreadsheet of ANS synaptic connectivity data values can be found in Supplementary file 1.
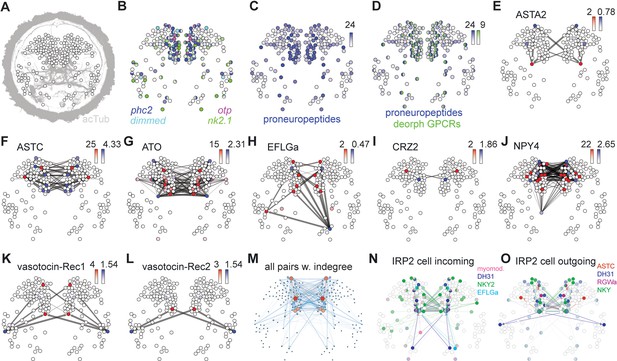
Mapping the peptidergic connectome in the Platynereis larval head.
(A) Positions of cells (nodes) from single cell RNA-Seq from 2-day-old larvae placed in an approximate spatial map, projected on an acetylated tubulin immunostaining (grey, anterior view). Samples with predicted bilateral symmetry were represented as two mirror-image nodes in the map. (B) Expression of neuroendocrine marker genes projected on the single-cell map. Color intensity of nodes reflects magnitude of normalized log10 gene expression. (C) Map of combined expression of 80 proneuropeptides expressed in each single-cell sample. (D) Map of combined expression of 80 proneuropeptides and 23 deorphanized GPCRs expressed in each single-cell sample. (E–L) Connectivity maps of individual neuropeptide-GPCR pairs, colored by weighted in-degree (red) and proneuropeptide log10 normalized expression (blue). Arrows indicate direction of signaling. Arrow thickness determined by geometric mean of log10 normalized proneuropeptide expression of signaling cell and log10 normalized GPCR expression of corresponding receiving cell. (M) Connectivity map of all possible known neuropeptide-GPCR signaling, color and node size represent weighted in-degree. (N) Connectivity map of an IRP2-expressing cell with incoming neuropeptide signals. (O) Connectivity map of an IRP2-expressing cell with outgoing neuropeptide signals.
-
Figure 3—source data 1
Platynereis reference transcriptome version 2.
This transcriptome was generated as described in (Conzelmann et al., 2013a), but with additional paired-end RNA-Seq data from 6 day old Platynereis, sequenced with an Illumina HiSeq 2000 (fasta file).
- https://doi.org/10.7554/eLife.26349.026
-
Figure 3—source data 2
Normalized transformed read counts for scRNA-seq data from Achim et al. (2015) mapped to our Platynereis reference transcriptome.
Includes spatial prediction from Achim et al. (2015): N = no prediction, Y1 = asymmetric cell predicted, Y2 = bilaterally symmetric pair of cells predicted.
- https://doi.org/10.7554/eLife.26349.027
-
Figure 3—source data 3
All-against-all pairwise correlation coefficients for normalized transformed read counts of scRNA-seq data from Achim et al. (2015) mapped to our Platynereis reference transcriptome.
This data was used to merge scRNA-seq samples that likely represented the same cell sourced from different larvae (see Materials and methods).
- https://doi.org/10.7554/eLife.26349.028
-
Figure 3—source data 4
Gexf connectivity map files generated from scRNA-Seq data for each peptide-receptor pair, and for all peptides by all receptors.
- https://doi.org/10.7554/eLife.26349.029
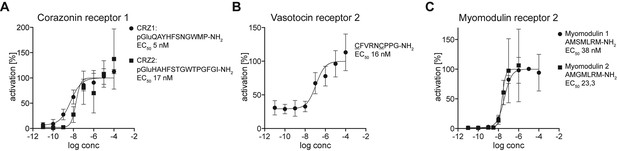
Dose-response curves of Platynereis deorphanized GPCRs treated with varying concentrations of peptides.
Data, representing luminescence units relative to the maximum of the fitted dose-response curves, are shown as mean ±SEM (n = 3). EC50 values and peptide sequences are shown beside each graph.
-
Figure 3—figure supplement 1—source data 1
Raw data (luminescence measurements) from deorphanization experiments.
- https://doi.org/10.7554/eLife.26349.018
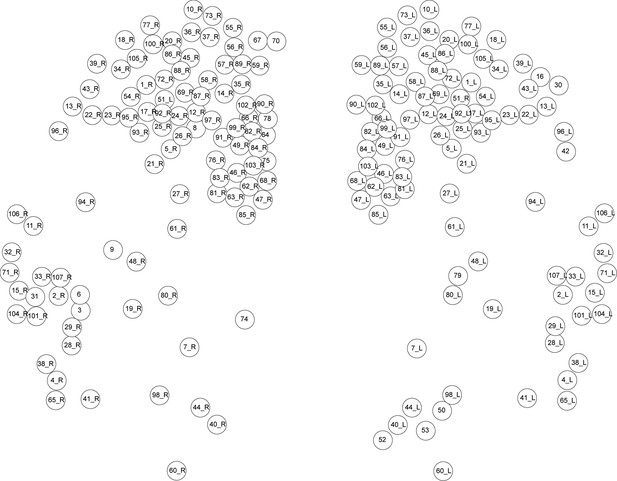
Spatial map of single cell RNA-Seq data from (Achim et al., 2015).
The position of each node on the map is an approximation based on the spatial predictions of each RNA-seq sample generated by Achim et al. from comparisons of transcriptome expression with a wholemount in situ hybridization gene expression atlas of 72 genes. The correspondence of node IDs to original sample IDs from Achim et al. (2015) is listed in Supplementary file 1.
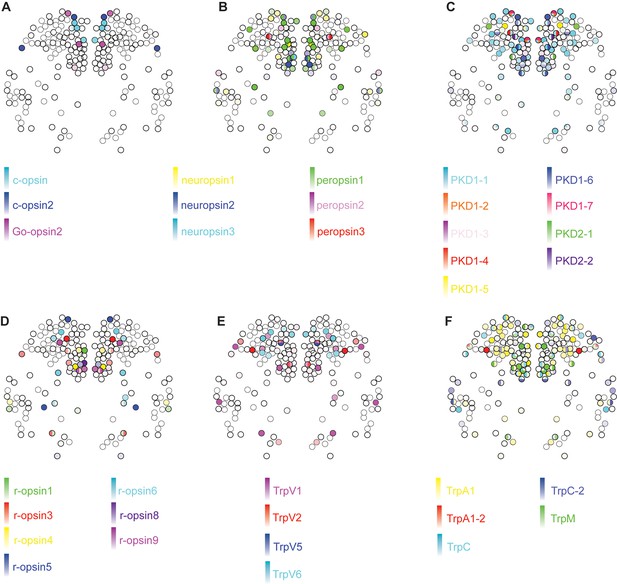
Expression of opsins and Trp channels in the Platynereis larval head.
Color intensity reflects relative normalized gene expression levels. Cells of the apical nervous system (i.e., cells expressing any combination of Phc2, dimmed, Otp and nk2.1) are indicated by a bold border.
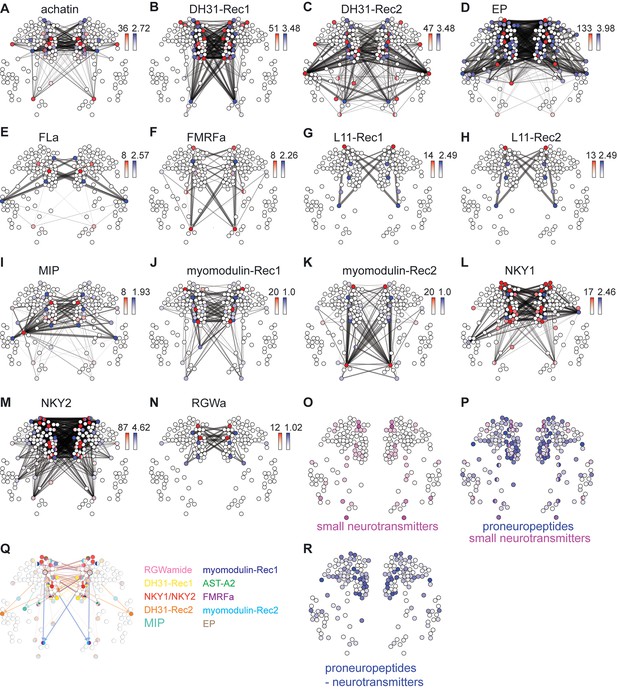
Neuropeptide-GPCR chemical connections in the Platynereis larval head.
(A–N) Connectivity maps of individual neuropeptide-GPCR pairs, colored by weighted in-degree (red) and proneuropeptide log10 normalized expression (blue). Arrows indicate direction of signaling. Arrow thickness determined by geometric mean of log10 normalized proneuropeptide expression of signaling cell and log10 normalized GPCR expression of corresponding receiving cell. (O) Expression map of small neurotransmitter synthesis markers. (P) Expression map of proneuropeptides and small neurotransmitter markers. (Q) Multichannel signaling from a highly peptidergic cell. (R) Expression map of proneuropeptides shown for those cells that do not express small neurotransmitter markers.
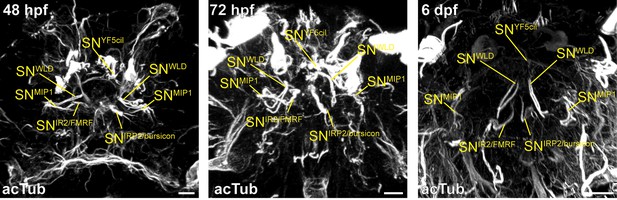
Comparison of spatial distribution of sensory cilia in the anterior head of 48 hpf, 72 hpf and 6 dpf Platynereis.
Cilia and the axonal scaffold of the larvae were labelled by immunostaining with an antibody raised against acetylated tubulin (white). Each sensory cilium is annotated according to its corresponding sensory neuron name in the reconstructed ssTEM data from a 72 hpf larva (yellow). Each staining can also be viewed in a 3D stack in the source data (tiff stacks). Scale bars: 10 µm.
-
Figure 3—figure supplement 5—source data 1
Tiff stacks of acetylated tubulin immunostaining from a 2-day-old, 3-day-old, and 6-day-old larva, anterior view.
- https://doi.org/10.7554/eLife.26349.023
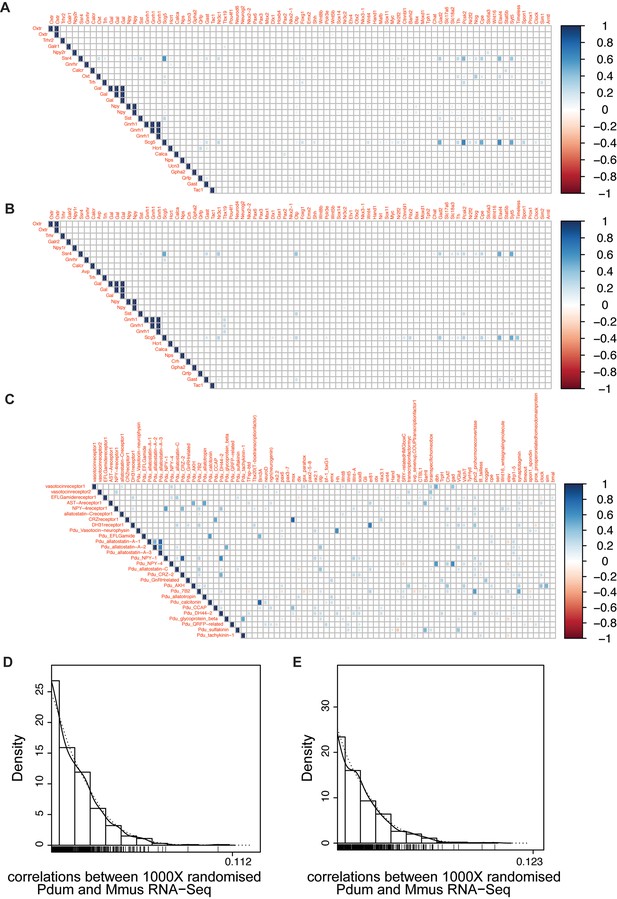
Comparison of gene co-expression correlations based on sc-RNA-Seq from Platynereis ANS and mouse hypothalamus datasets.
(A) Co-expression correlation heatmap for mouse data (Romanov et al., 2017 dataset). (B) Co-expression correlation heatmap for mouse data (Campbell et al., 2017 dataset). (C) Co-expression correlation heatmap for Platynereis data. (D and E) Distributions of correlations of co-expression values between Platynereis and mouse (Romanov et al., 2017) (D) and Campbell et al. (2017) (E) datasets after 1000X randomization. 0.112 and 0.123 are the global correlation values calculated on, respectively, the Romanov et al., 2017 and the Campbell et al. (2017) datasets, and rank second and first, respectively, compared to all other 1000 correlations computed on the randomized datasets from each of the two datasets.
-
Figure 3—figure supplement 6—source data 1
List of Platynereis and mouse gene orthologs ‘ortholog_table_pdum_mouse_clean.txt’; Global correlation of Platynereis and mouse orthologous marker gene data sets ‘correlation_pdum_mouse_romanov.txt’, ‘correlation_pdum_mouse_Campbell.txt’; Tables of correlation coefficients and p-values from comparison of Platynereis and mouse scRNA-Seq datasets, Achim et al. (2015) versus Romanov et al. 2016 ‘conserved_coexpression_pdum_Romanov.txt’, Achim et al. (2015) versus Campbell et al. (2017), ‘conserved_coexpression_pdum_campbell.txt’.
- https://doi.org/10.7554/eLife.26349.025
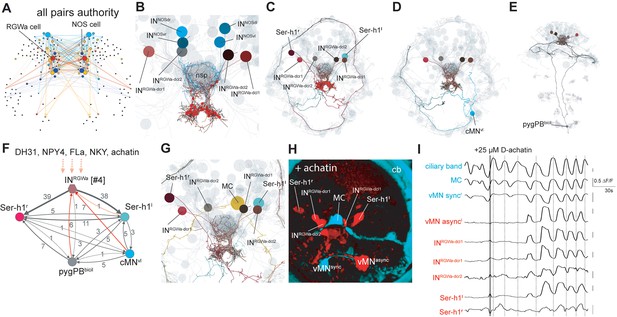
Projection neurons connect the ANS to the synaptic nervous system.
(A) Connectivity map of all possible known neuropeptide-GPCR signaling, colors represent modules defined by randomized community detection analysis in Gephi and node size represents authority. (B) Reconstructed INRGWaand INNOS projection neurons (ANS) (shades of brown and cyan) in a full body transmission electron microscopy (ssTEM) dataset of a 3-day-old larva, shown against a framework of ANS neurons (light grey). Axons and dendrites appear as lines and cell body positions are represented by spheres. (C) ssTEM reconstruction of INRGWa projection neurons and Ser-h1 serotonergic neurons. Red spheres indicate INRGWa presynaptic sites. (D) ssTEM reconstruction of INRGWa projection neurons and a presynaptic ciliomotor neuron, cMNvl. (E) ssTEM reconstruction of INRGWa neurons and a presynaptic sensory neuron, pygPDbicil. (F) Synaptic connectivity graph of INRGWa, Ser-h1, pygPDbicil, and cMNvl neurons. Synaptic inputs from the synaptic nervous system to the INRGWa neuron are in red. Peptidergic inputs to the INRGWa neurons are indicated with dashed arrows. (G) ssTEM reconstruction of INRGWa projection neurons, Ser-h1 neurons, and the cholinergic ciliomotor MC neuron, anterior view (H) Confocal microscopy image of correlated pixels of GCaMP6s signal in a 2-day-old Platynereis larva after the addition of 25 µM D-achatin neuropeptide, anterior view. Cells showing correlated activity with the serotonergic neurons (red) and the MC cell (cyan) are shown. INRGWa-dcl2 could not be identified in this larva and is likely obscured by the MC cell, Ser-h1l and/or INRGWa-dcl1. (I) Neuronal activity patterns of individually identified neurons in a 2-day-old larva treated with 25 µM achatin.
-
Figure 4—source data 1
Video of calcium imaging in a Platynereis larva used to calculate neuronal activity correlations in Figure 4H and neuronal activity patterns in Figure 4I (.tiff file).
- https://doi.org/10.7554/eLife.26349.034
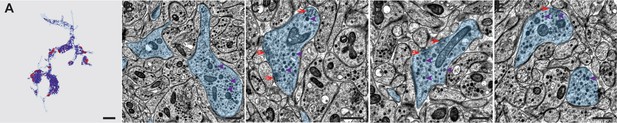
Spatial distribution of dense core vesicles and synapses in an RGWa-expressing projection interneuron, INRGW-dcr1.
(A) Reconstruction of 50 layers of TEM sections from the 72 hpf larva ssTEM dataset containing the axon of INRGWa-dcr1 in the larval neurosecretory plexus. The axon is coloured light blue, synaptic sites are red, and dense core vesicles purple. Scale bar = 2 µm. This reconstruction can be viewed in 3D in Video 3. (B–E) TEM sections from different z-positions in the 72 hpf larva ssTEM dataset containing the neurites of the INRGWa-dcr1 axon (blue). Examples of dense core vesicles are indicated by purple arrowheads, red arrows indicate synapses (clusters of small clear vesicles at the neurite border). Scale bar A: 2 µm; scale bar B-E: 500 nm.
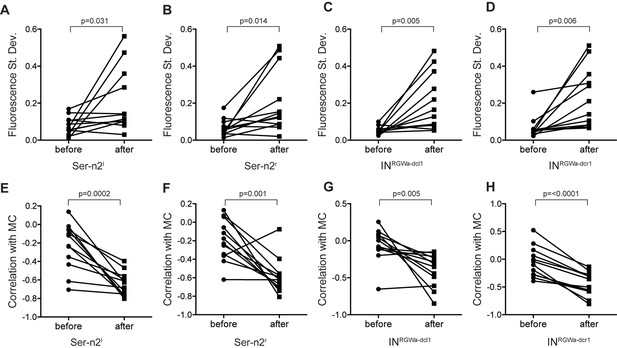
(A–D) Standard deviation in fluorescent calcium signal of the (A) Ser-h1l, (B) Ser-h1r, (C) INRGWa-dcl1 and (D) INRGWa-dcr1 neurons in 2-day-old larvae before and after treatment with 25 µM D-achatin.
(E–H) Correlation of neuronal calcium signals of (E) Ser-h1l, (F) Ser-h1r, (G) INRGWa-dcl1 and (H) INRGWa-dcr1 with calcium signals measured from the MC cell in 2-day-old larvae before and after treatment with 25 µM D-achatin.
-
Figure 4—figure supplement 2—source data 1
Correlation values of neuronal activity patterns and standard deviation of GCaMP6 fluorescence in achatin-treated larvae.
- https://doi.org/10.7554/eLife.26349.033
Videos
Anterior view of the Platynereis 3 day ANS reconstructed from ssTEM data.
Circles represent cell bodies and lines are axons and dendrites. ANS neurons (coloured) appear against a backdrop of neurons from the eye photoreceptor cells (dark blue-grey) and ciliated cells of the prototroch, and anterior crescent cell (light grey). The axons of the ANS neurons project to the center of the larval head, where they form a dense spherical plexus that sits anterior to the rest of the larval nervous system.
Blender model of central sensory neurons of the ANS reconstructed from ssTEM data, anterior view.
Only the cell body, dendrite and sensory cilia of each cell were reconstructed. Includes the SNIRP2, SNMIP1, SNWLD and SNYF5cil neurons. These neurons could be assigned molecular identities based on comparison of their morphology in the TEM data reconstructions to morphological data from immunostainings and in situ hybridizations counterstained with acetylated tubulin antibody (Figure 1—figure supplements 4 and 5).
3D reconstruction from 50 TEM sections of a segment of the axon of the projection interneuron INRGWa-dcr1, to show distribution of dense core vesicles and classical synapses.
The axon is coloured light blue, synaptic sites are red, and dense core vesicles purple. Scale bar = 2 µm.
Additional files
-
Supplementary file 1
.xls file containing ANS synaptic connectivity spreadsheet, node ID to cell ID key, chemical network parameters, and log10 normalized expression values from mapping of single cell data for neuropeptides, GPCRs, sensory genes and neurotransmitter synthesis enzymes in separate worksheets.
- https://doi.org/10.7554/eLife.26349.036
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26349.037





