Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells
Figures
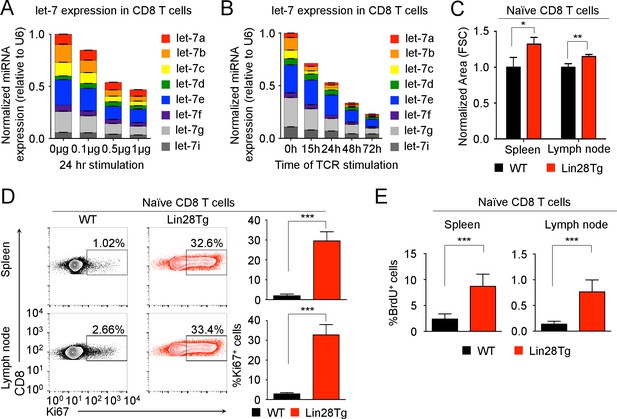
let-7 expression is necessary and sufficient to maintain the naive phenotype of CD8 T cells prior to TCR stimulation.
(A) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive CD8 T cells stimulated with plate-bound anti-TCR (μg as indicated) and anti-CD28 (5 μg) for 24 hr, presented relative to results obtained for the small nuclear RNA (U6 control) and normalized to the unstimulated (0 μg) sample. (B) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive CD8 T cells stimulated with plate-bound anti-TCR (5 μg) and anti-CD28 (5 μg) over increasing periods of time as indicated, presented relative to results obtained for the small nuclear RNA (U6 control) and normalized to the unstimulated (0 hr) sample. (C) Size analysis based on FSC (forward scatter) of naive CD8 T cells from the spleens and lymph nodes of P14+wild-type and P14+Lin28Tg mice, both on Rag2−/− background, normalized to wild-type. (D) Expression of Ki67 in naive CD8 T cells from spleens and lymph nodes of P14+wild-type and P14+Lin28Tg mice, both on Rag2−/− background (left). Quantification of the frequency of Ki67+ cells in these populations (right). (E) Frequency of BrdU+ cells in naive CD8 T cells from spleens and lymph nodes of P14+wild-type (n = 6) and P14+Lin28Tg (n = 5) mice, both on Rag2−/− background, labeled with BrdU in vivo. *p<0.05, **p<0.01,***p<0.001, compared with wild-type using two-tailed Student’s t-test. Data are one experiment representative of three independent experiments (a, b; mean and s.e.m. of technical triplicates; c, d; mean and s.e.m. of three experiments).
-
Figure 1—source data 1
Quantification of the expression of miRNAs by qPCR, and flow cytometry analysis of the phenotype of naive CD8 T cells.
- https://doi.org/10.7554/eLife.26398.003
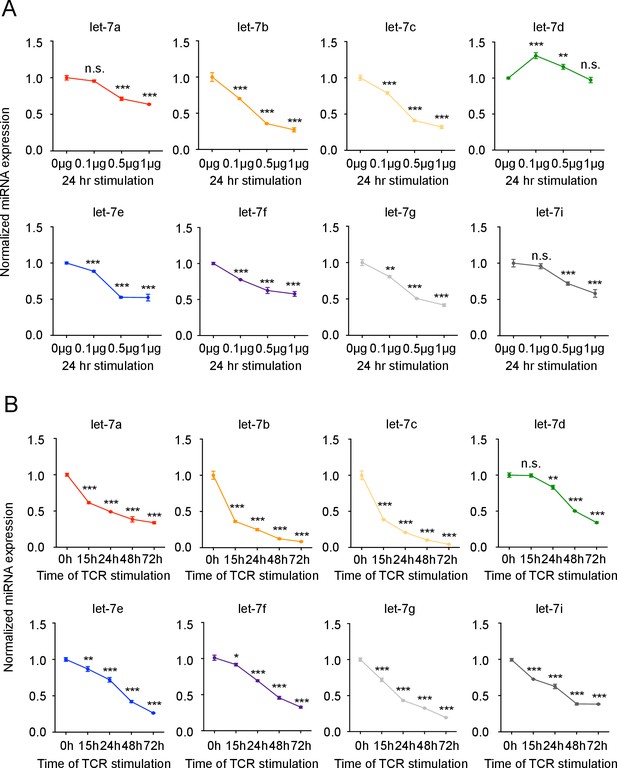
let-7 expression is downregulated upon TCR stimulation in CD8 T cells.
(A) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive CD8 T cells stimulated with plate-bound anti-TCRβ (μg as indicated) and anti-CD28 (5 μg) for 24 hr (from Figure 1A), presented relative to results obtained for the small nuclear RNA (U6 control) and normalized to the unstimulated (0 μg) sample, and displayed as individual miRNAs. (B) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive CD8 T cells stimulated with plate-bound anti-TCRβ (5 μg) and anti-CD28 (5 μg) over increasing periods of time as indicated (from Figure 1B), presented relative to U6 control and normalized to the unstimulated (0 hr) sample, and displayed as individual miRNAs. n.s., not significant (p>0.05), *p<0.05, **p<0.01, and ***p<0.001, compared with 0 μg (A) or 0 hr (B) using two-tailed Student’s t-test (A, B). Data are one experiment representative of three independent experiments (b; mean and s.e.m. of technical triplicates).
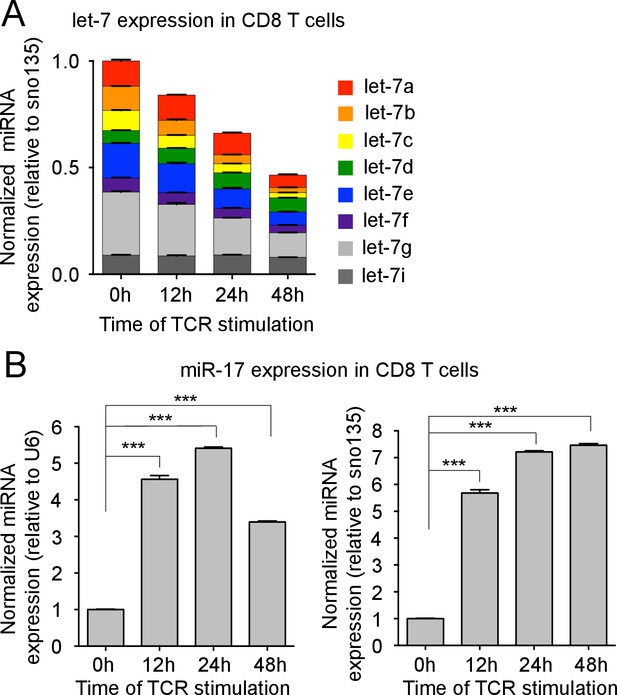
TCR-mediated regulation of miRNA expression.
(A) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive CD8 T cells stimulated with plate-bound anti-TCRβ (10 μg) and anti-CD28 (5 μg) over increasing periods of time as indicated, presented relative to results obtained for the small nucleolar RNA (sno135) and normalized to the unstimulated (0 hr) sample. (B) Quantitative RT-PCR analysis of miR-17 in naive CD8 T cells stimulated with plate-bound anti-TCRβ (10 μg) and anti-CD28 (5 μg) over increasing periods of time as indicated, presented relative to results obtained for the small nuclear RNA (U6) (left) or the small nucleolar RNA (sno135) (right) and normalized to the unstimulated (0 hr) sample. ***p<0.001, compared with 0 hr using two-tailed Student’s t-test (B). Data are from one experiment representative of two independent experiments (a, b; mean and s.e.m. of technical triplicates).
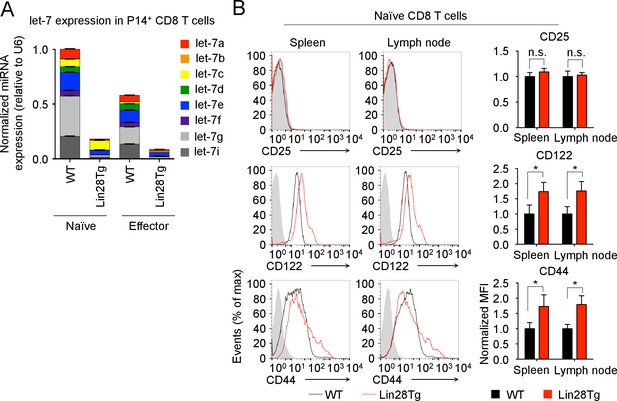
Expression of let-7 and activation markers in P14+ CD8 T cells.
(A) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive and effector (day 5) TCR transgenic CD8 T cells from P14+Rag2−/−, and P14+Lin28TgRag2-/− mice. (B) Surface expression (right) and normalized MFI (left) of CD25, CD44 and CD122 on naive CD8 T cells from both spleens and lymph nodes of P14+wild-type and P14+ Lin28Tg mice, both on Rag2−/− background. Gray indicates an isotype control for staining. n.s., not significant (p>0.05), *p<0.05, **p<0.01, and ***p<0.001, compared with wild-type using two-tailed Student’s t-test (B). Data are from one experiment representative of three independent experiments (a; mean and s.e.m. of technical triplicates; b, c; mean and s.e.m. of three experiments).
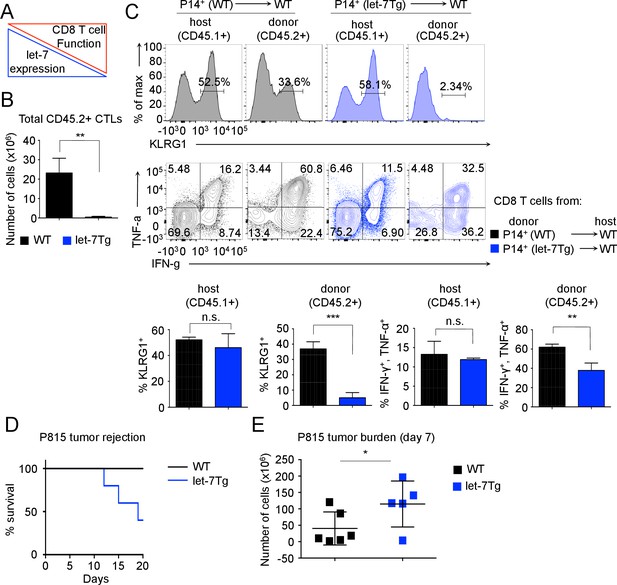
Inhibitory role of let-7 miRNA expression in CD8 T-cell-mediated responses in vivo.
(A) Schematic representation of the hypothesis that let-7 expression inhibits the differentiation of CD8 T cells. (B) Quantification of the number of donor (CD45.2+) CD8 T cells from P14+wild-type (n = 3) or P14+let-7Tg mice (n = 3) in the spleens of congenic (CD45.1+) host mice 7 days after cell transfer and LCMV Armstrong infection. (C) Surface expression of the activation marker KLRG1 on wild-type host, and indicated donor cells (top). Expression of IFN-γ, and TNF-α in wild-type host, and donor LCMV-specific CD8 T cells from the indicated mice, as determined by re-stimulation with the gp33 peptide, and subsequent intracellular staining (middle). Quantification of the frequency of KLRG1+, and IFN-γ+, TNF-α+ populations in wild-type host, and donor cells from the indicated mice (bottom). (D) Analysis of the survival of wild-type (n = 5) or let-7Tg (n = 5) mice injected i.p. with 30 × 106 P815 cells. (E) Quantification of the number of P815 tumor cells remaining in the peritoneal cavity 7 days after i.p. injection of 20 × 106 cells into either wild-type (n = 6), or let-7Tg mice (n = 5). n.s., not significant (p>0.05), *p<0.05, **p<0.01, and ***p<0.001, compared with wild-type using two-tailed Student’s t-test (B, C, D) or one-tailed Student’s t-test (E). Data are from one experiment representative of three independent experiments (b, c; mean and s.e.m. of technical triplicates), or two experiments (D,E).
-
Figure 2—source data 1
Quantification of let-7 expression by qPCR, and flow cytometry analysis of effector CD8 T cells during the response to LCMV infection.
- https://doi.org/10.7554/eLife.26398.008
P815 (H-2d) tumor rejection is mediated by CD8 T cells in H-2b mice.
(A) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive and effector (day 5) TCR transgenic CD8 T cells from P14+Rag2−/−, and P14+let-7TgRag2−/− mice (B) Analysis of the survival of Rag2−/− mice injected i.p. with 30 × 106 P815 cells, which received i.v. adoptive transfer of 10 × 106 naïve purified CD8 T cells (n = 6) or no T cells at all (n = 8). Data are from one experiment representative of three independent experiments (a; mean and s.e.m. of technical triplicates) or one experiment (B).
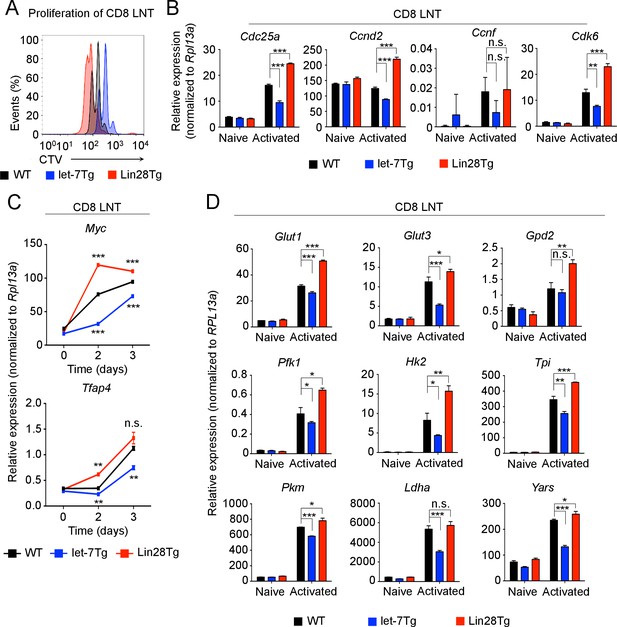
let-7 miRNAs suppress the proliferation and metabolism of activated CD8 T cells.
(A) Analysis of the proliferation of CellTrace Violet-labeled naive CD8 T cells from the indicated mice 72 hr after activation with anti-CD3 mAbs. (B) Quantitative RT-PCR analysis of cell cycle regulators: Cdc25a (Cell division cycle 25A phosphatase), Ccnd2 (Cyclin D2), Ccnf (Cyclin F), Cdk6 (Cyclin dependent kinase 6) in naive and activated wild-type, let-7Tg, and Lin28Tg CD8 T cells 3 days after anti-CD3 mAb stimulation, presented relative to the ribosomal protein Rpl13a. (C) Quantitative RT-PCR analysis of Myc and Tfap4 (Transcription factor AP-4) in CD8 T cells after stimulation with anti-CD3 mAbs, presented relative to the ribosomal protein Rpl13a. Wild-type, let-7Tg, and Lin28Tg CD8 T cells were stimulated with anti-CD3 mAbs and differentiated for the indicated time. (D) Quantitative RT-PCR analysis of the expression of genes involved in the metabolic switch: Glut1 (Glucose transporter 1), Glut3 (Glucose transporter 3), Gpd2 (Glycerol-3-phosphate dehydrogenase 2), Pfk1 (Phosphofructokinase 1), Hk2 (Hexokinase 2), Tpi (Triose phosphate isomerase), Pkm (Pyruvate kinase muscle isozyme), Ldha (Lactate dehydrogenase A), Yars (Tyrosyl-tRNA synthetase) in wild-type, let-7Tg, and Lin28Tg CD8 T cells 3 days after stimulation with anti-CD3 mAbs, presented relative to the ribosomal protein, Rpl13a. n.s., not significant (p>0.05), *p<0.05, **p<0.01, and ***p<0.001, compared with wild-type using two-tailed Student’s t-test. (a, b, c, d; one experiment representative of three independent experiments (a) or mean and s.e.m. of technical triplicates (b, c, d)).
-
Figure 3—source data 1
Quantification of the expression of cell cycle regulators, Myc and Tfap4, metabolic enzymes, and let-7 miRNAs by qPCR.
- https://doi.org/10.7554/eLife.26398.011
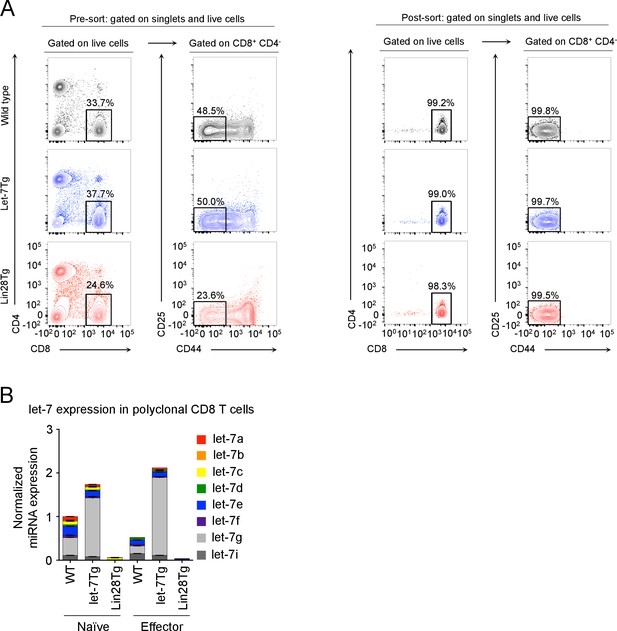
Sorting strategy and let-7 expression in polyclonal CD8 T cells.
(A) Pre- sort and post- sort purities for naive CD8 T cells from wild-type, let-7Tg, and Lin28Tg mice. Cells were isolated from the lymph nodes of the indicated mice, and enriched for T cells using anti-mouse IgG magnetic beads. Next, to acquire naive CD8 T cells, lymph node T cells were then stained for CD25, CD44, CD4, and CD8, and sorted as the CD25-CD44loCD4-CD8+ population. (B) Quantitative RT-PCR analysis of individual let-7 miRNAs in naive and activated (day 5) polyclonal CD8 T cells from wild-type, let-7Tg, and Lin28Tg mice.
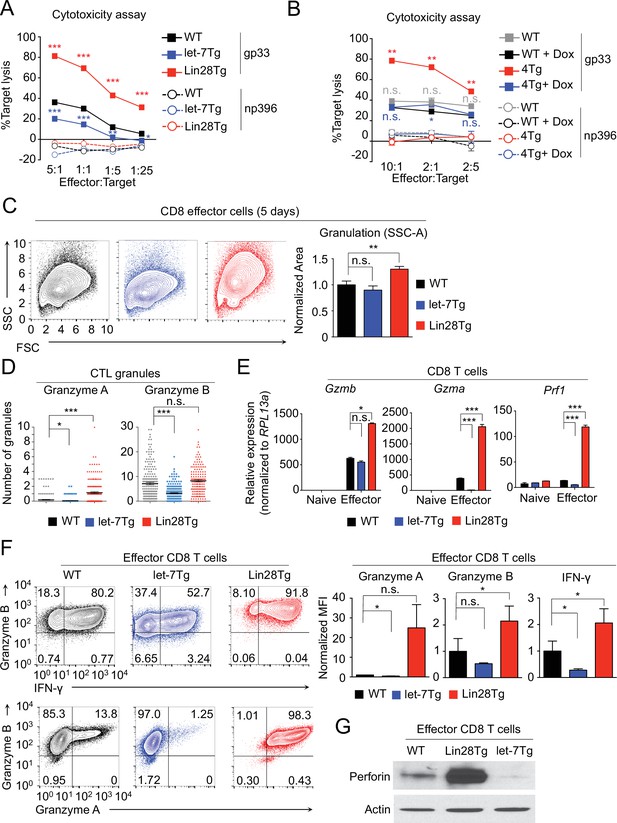
let-7 miRNAs negatively regulate differentiation and acquisition of effector function in CTLs.
(A, B) Cytotoxicity assay of differentiated CTLs from P14+wild type, P14+let-7Tg, and P14+Lin28Tg lymph nodes (A) or from P14+wild-type (WT), or P14+let-7Tg+Lin28Tg+Rag2−/− (4Tg) lymphocytes co-cultured with either LCMV gp33 or LCMV np396 peptide-pulsed splenocytes for 4–5 hr, either in the presence or absence of doxycycline (B). (C) Analysis of the internal complexity (FSC, forward scatter; SSC, side scatter) of effector (5 days after anti-CD3 mAb stimulation) CD8 T cells generated from wildtype, let-7Tg, and Lin28Tg lymphocytes (left) and quantification of SSC of CD8 T cells normalized to wild-type. (D) Quantification of Granzyme A and Granzyme B- positive granules in wild-type, let-7Tg, and Lin28Tg CTLs via MilliPore Amnis ImageStream. (E) Quantitative RT-PCR analysis of effector molecule mRNA expression: Gzma (Granzyme A), Gzmb (Granzyme B), Prf1 (Perforin) in naive and effector CD8 T cells from wild-type, let-7Tg, and Lin28Tg lymph nodes, presented relative to expression of the ribosomal protein Rpl13a. (F) Staining (top, middle) and MFI (bottom) of Granzyme B, Interferon-γ, and Granzyme A in wild-type, let-7Tg, and Lin28Tg effector CD8 T cells normalized to wild type. (G) Western blot analysis of lysates of wild-type, let-7Tg, and Lin28Tg effector CD8 T cells, probed with monoclonal antibodies against Perforin and actin. n.s., not significant (p>0.05), *p<0.05, **p<0.01, and ***p<0.001, compared with wild-type using two-tailed Student’s t-test. Data are from one experiment representative of three experiments (a, b, e; mean and s.e.m. of technical triplicates, f; mean and s.e.m of three experiments).
-
Figure 4—source data 1
Quantification of cytotoxic assays and expression of effector molecules assessed by qPCR and flow cytometry in CTLs.
- https://doi.org/10.7554/eLife.26398.014
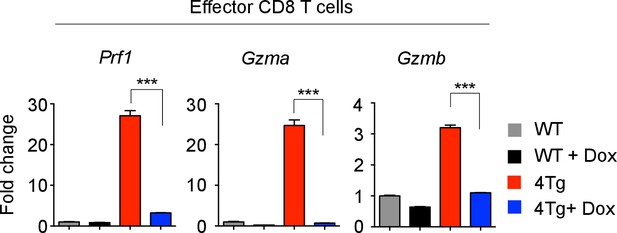
Effector molecule mRNA expression in 4Tg mice.
Quantitative RT-PCR analysis of effector molecule mRNA expression: Prf1 (Perforin), Gzma (Granzyme A), Gzmb (Granzyme B) in effector CD8 T cells from P14+wild-type, or P14+let-7Tg+Lin28Tg+Rag2−/− (4Tg) lymph nodes cultured either in the presence or absence of Dox (doxycycline), presented as the fold change in expression, normalized to wild-type. ***p<0.001, 4Tg without Dox compared with 4Tg with Dox using two-tailed Student’s t-test. Data are from one experiment (mean and s.e.m. of technical triplicates).
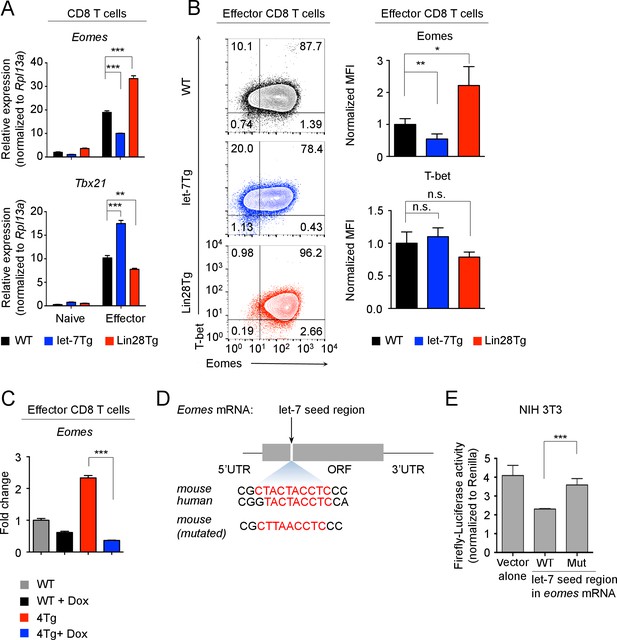
let-7 miRNAs directly target the mRNA of Eomes in activated CD8 T cells.
(A) Quantitative RT-PCR analysis of Eomes (Eomesodermin) and Tbx21 (T-bet) mRNA in naive and effector CD8 T cells (5 days after anti-CD3 mAb stimulation) generated from wild-type, let-7Tg, and Lin28Tg lymphocytes, presented relative to the ribosomal protein Rpl13a. (B) Staining of Eomes and T-bet (left) and MFI of Eomes and T-bet (right) in wild-type, let-7Tg and Lin28Tg effector CD8 T cells, normalized to wild-type. (C) Quantitative RT-PCR analysis of Eomes (Eomesodermin) mRNA in P14+wild-type, or P14+let-7Tg+Lin28Tg+Rag2−/− (4Tg) lymph nodes cultured either in the presence or absence of Dox, presented as the fold change in expression, normalized to wild-type. (D) Eomes mRNA, including the 5’ and 3’ untranslated regions (UTR) and the open-reading frame (ORF) within which one let-7-binding site was identified (top), sequence conservation between the mouse and human let-7-binding sites of Eomes is shown (middle). Mutated sequence of the let-7-binding site in the cDNA of mouse Eomes (bottom) used in the luciferase reporter assay. (E) Luciferase reporter assay of let-7 targeting of the Eomes ORF in NIH 3T3 fibroblasts transfected with a luciferase reporter containing either the intact or mutated sequence of the let-7 seed region from the mouse Eomes ORF; the activity of firefly luciferase was normalized to the Renilla luciferase control. n.s., not significant (p>0.05), *p<0.05, **p<0.01, and ***p<0.001, compared with wild-type using two-tailed Student’s t-test. Data are from one experiment representative of three independent experiments (a, b, c, e; mean and s.e.m. of technical triplicates (A,C,E) or three experiments (B)).
-
Figure 5—source data 1
Quantification of luciferase activity, and the expression of transcription factors in CTLs assessed by flow cytometry and qPCR.
- https://doi.org/10.7554/eLife.26398.017
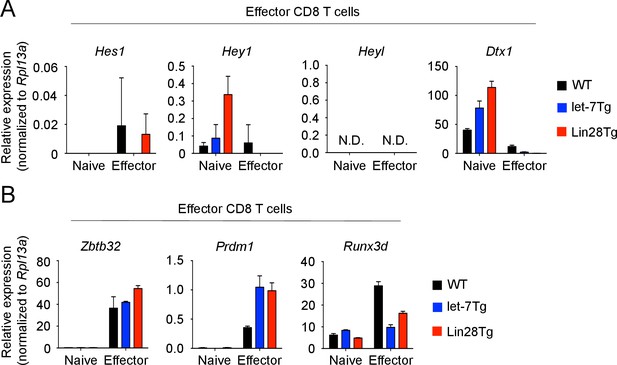
Effect of let-7 miRNAs on expression of transcription factors in effector CD8 T cells.
(A, B) Quantitative RT-PCR analysis of Notch target genes: Hes1 (Hes family BHLH transcription factor 1), Hey1 (Hes related family BHLH transcription factor with YRPW motif 1), Heyl (Hes related family BHLH transcription factor with YRPW motif- like), Dtx1 (Deltex 1) (A) and transcription factors that control CD8 T cell differentiation: Zbtb32 (Zinc finger and BTB domain containing 32), Prdm1 (Blimp-1), Runx3d (Runt related transcription factor three distal) (B) in naive and effector (5 days after anti-CD3 mAb stimulation) wild-type, let-7Tg, and Lin28Tg CD8 T cells, presented relative to the expression of the ribosomal protein Rpl13a. Data are from one experiment representative of three independent experiments (a, b; mean and s.e.m. of technical triplicates). N.D., not determined.
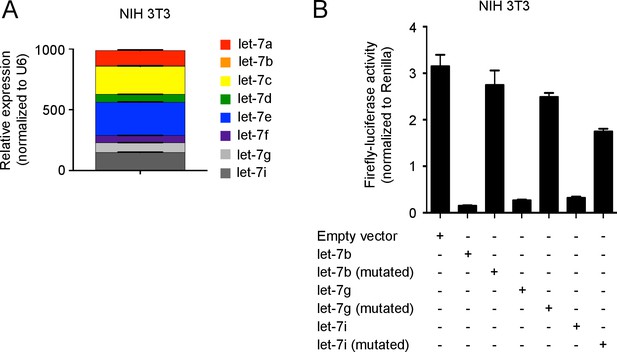
Let-7 expression and luciferase detection in NIH 3T3 fibroblasts.
(A) Quantitative RT-PCR analysis of individual let-7 miRNAs in NIH 3T3 fibroblasts, presented relative to results obtained for the small nuclear RNA (U6) (B) Expression analysis of three let-7 family members by luciferase reporter assay. A firefly luciferase reporter containing either intact or mutated let-7b, let-7g or let-7i antisense seed regions were transfected into NIH 3T3 cells. The activity of firefly luciferase was normalized to the Renilla luciferase control. Data are one experiment representative of three independent experiments (a, b; mean and s.e.m. of technical triplicates).
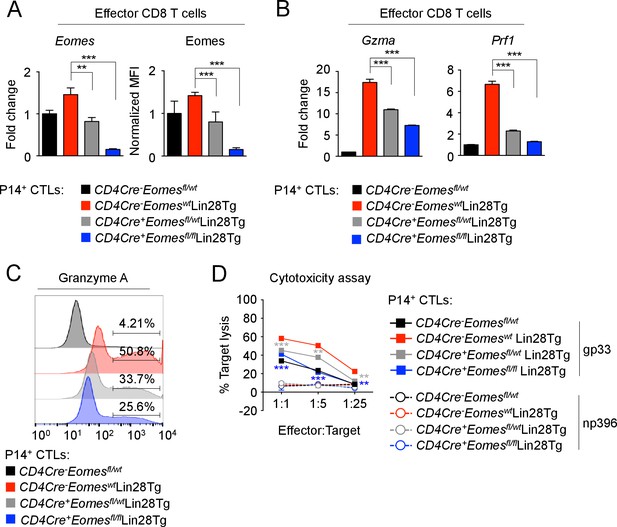
let-7 miRNAs control the differentiation of CTLs through Eomes-dependent and independent mechanisms.
(A) Quantitative RT-PCR analysis of Eomes mRNA, presented as the fold change in expression, normalized to wild-type (left) and MFI of Eomes protein expression, normalized to wild-type (right), from CTLs generated from P14+CD4Cre-Eomesfl/wt, P14+CD4Cre-EomeswtLin28Tg, P14+CD4Cre+Eomesflfl/wtLin28Tg, and P14+CD4Cre+Eomesflfl/flLin28Tg CD8 T cells. (B) Quantitative RT-PCR analysis of GzmA (Granzyme A) and Prf1 (Perforin) mRNA in effector CTLs generated from the indicated mice, presented as the fold change in expression, normalized to wild-type. (C) Staining of Granzyme A in CTLs generated from the indicated mice. (D) Cytotoxicity assay demonstrating specific target lysis of differentiated P14+ CTLs from the indicated mice, co-cultured with either LCMV gp33 or LCMV np396 peptide-pulsed splenocytes for 4–5 hr. **p<0.01, and ***p<0.001, compared with Lin28Tg using two-tailed Student’s t-test. Data are from one experiment representative of two independent experiments (a; mean and s.e.m. of two experiments; b, d; mean and s.e.m. or technical triplicates).
-
Figure 6—source data 1
Quantification of cytotoxic assays and expression of effector molecules assessed by qPCR and flow cytometry in CTLs.
- https://doi.org/10.7554/eLife.26398.021
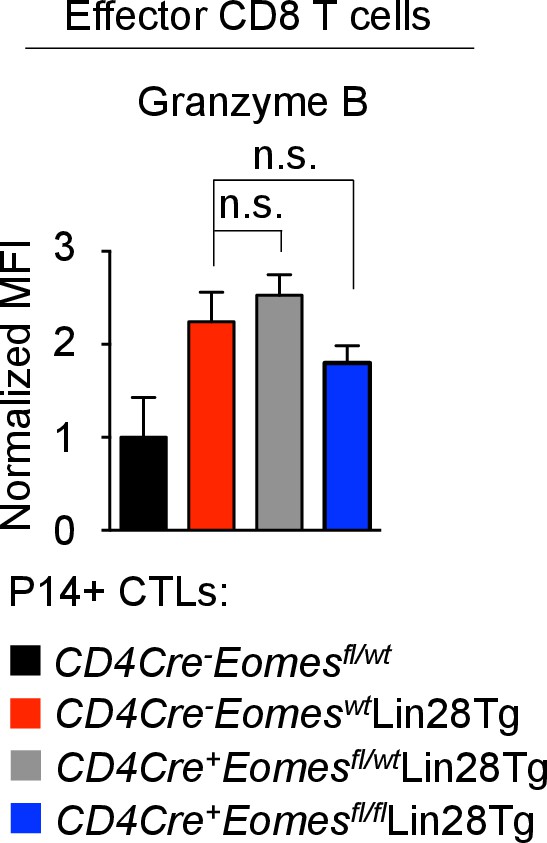
Granzyme B expression is regulated independent of Eomes.
Staining of Granzyme B in CTLs generated from the indicated mice. n.s., not significant (p>0.05), 4Tg without Dox compared with 4Tg with Dox using two-tailed Student’s t-test. Data are from one experiment representative of two independent experiments (mean and s.e.m. of two experiments).
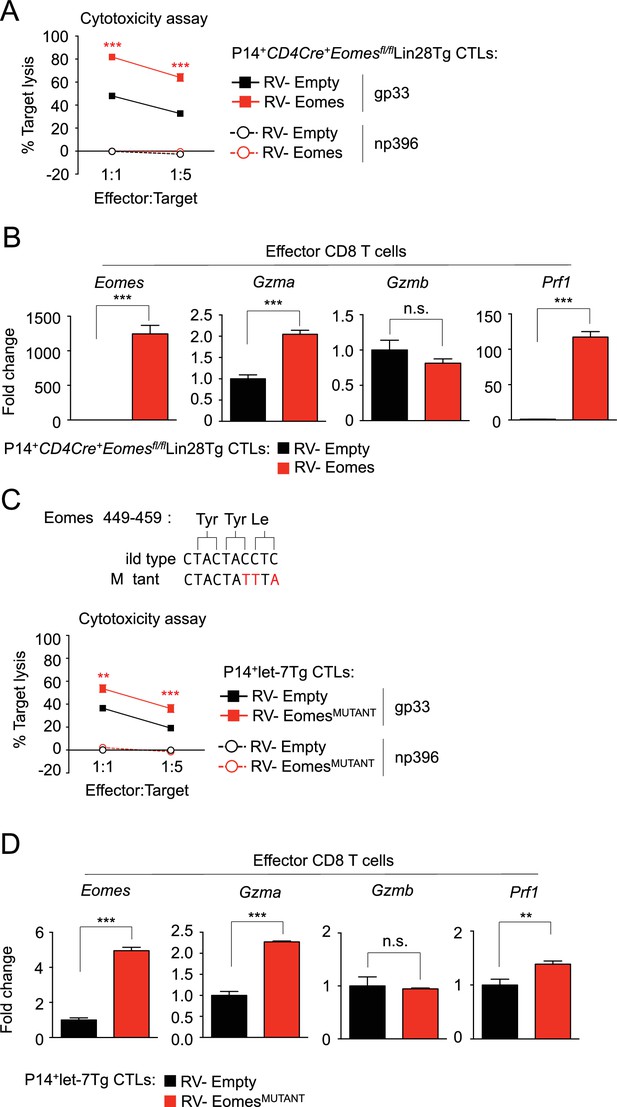
Re-expression of Eomes enhances cytotoxic function of CTLs.
(A) Cytotoxicity assay demonstrating specific target lysis of LCMV gp33 or LCMV np396 peptide-pulsed splenocytes by differentiated P14+CD4Cre+Eomesflfl/flLin28Tg CTLs transduced with the indicated virus, cells were co-cultured for 4–5 hr. (B) Quantitative RT-PCR analysis of Eomes (Eomesodermin), Gzma (Granzyme A), Gzmb (Granzyme B), and Prf1 (Perforin) expression in CTLs described in a, presented as the fold change in expression, normalized to RV-Empty. (C) Visualization of the mutations made to the let-7-binding motif in mouse Eomes ORF to generate the RV-EomesMUTANT (top), and cytotoxicity assay demonstrating specific target lysis of differentiated P14+let-7Tg CTLs transduced with the indicated virus, co-cultured with either LCMV gp33 or LCMV np396 peptide-pulsed splenocytes for 4–5 hr (bottom). (D) Quantitative RT-PCR analysis of Eomes (Eomesodermin), Gzma (Granzyme A), Gzmb (Granzyme B), and Prf1 (Perforin) in effector CTLs from c, presented as the fold change in expression, normalized to RV-Empty. n.s., not significant (p>0.05), **p<0.01, ***p<0.001, RV-Eomes or RV-EomesMUTANT compared with RV-Empty using two-tailed Student’s t-test (A, B, C, D). Data are from one experiment representative of two independent experiments (a, b, c, d; mean and s.e.m. of technical triplicates).
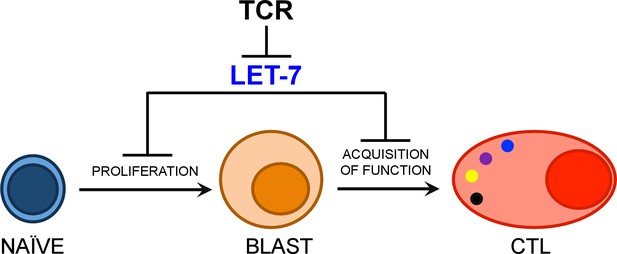
Model of let-7- mediated regulation of CD8 T cell differentiation.
Upon TCR stimulation, let-7 miRNAs are rapidly down-regulated, in order to release the ‘molecular brake’ let-7 places on CD8 T cell differentiation in naive cells. Our study suggests that let-7 miRNAs inhibit clonal expansion and metabolic reprogramming of activated CD8 T cells through inhibition of Myc and other cell cycle controlling factors. Further, our data indicates let-7 also suppresses the acquisition of effector function of CTLs through the direct targeting of Eomes mRNA.
Additional files
-
Supplementary file 1
A list of all genes containing a 9-base pair let-7 binding motif in either the 3’ untranslanted region (UTR) or open reading frame (ORF) from both the mouse and human genomes, where a (+) or (-) demonstrates the orientation of the gene in the genome.
The ending models are an illustration of the location of the let-7 binding motifs within the human and mouse Eomes gene, where red represents exons and pink represents introns.
- https://doi.org/10.7554/eLife.26398.025
-
Supplementary file 2
Primer sequences and Taqman Assay catalog numbers.
- https://doi.org/10.7554/eLife.26398.026





