ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate
Figures
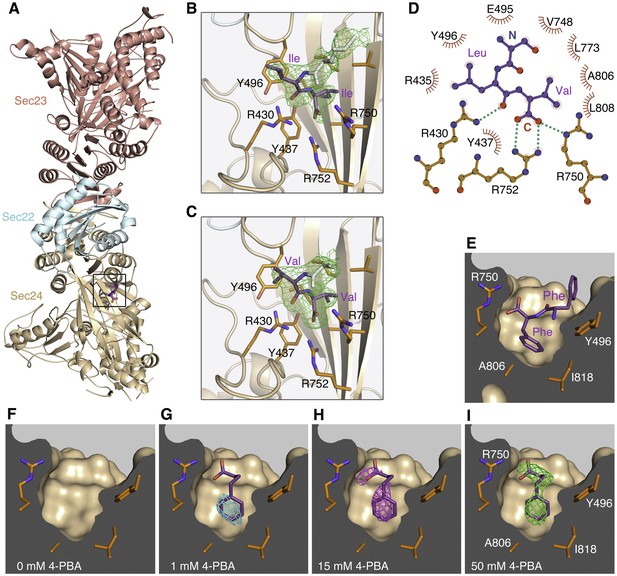
COPII binding site for the ΦC signal motif and 4-PBA located by X-ray crystallography.
(A) Crystal structure of the COPII•SNARE complex comprising human Sec23a/Sec24a•Sec22b bound to a ΦC peptide (boxed). The concave, membrane-binding surface of the coat faces right. (B) Close-up view of the B site on Sec24a with bound ΦC sequence containing the terminal Ile-Ile motif (colored purple) of the S. cerevisiae p24 protein Erv25p. Green contour lines show residual electron density (Fo-Fc synthesis with no phase bias) at 2.6 Å resolution, contoured at 2.6 σ. Electron density is also observable for N-terminal residues of the peptide (residues colored light grey). Key side chains of Sec24 are labeled (colored orange). See Table 1 for details of peptide-bound crystal structures. (C) Residual (Fo-Fc) electron density (2.6 Å resolution, 2.6 σ contour) for a bound ΦC peptide containing the terminal Val-Val motif of human p24β1. (D) Schematic diagram shows the bonding arrangement to the potent ΦC motif Leu-Val. Diagram is based on the structure of Sec23a/Sec24a•Sec22b bound to the sequence EVTSLV (see Table 1). The label N denotes the serine residue upstream of the Leu-Val motif. (E) Conformation of a ΦC peptide containing the terminal Phe-Phe motif of ERGIC-53 bound to Sec24a (drawn as a surface representation). (F–I) Residual (Fo-Fc) electron density (2.8 Å resolution, 2.3 σ contour, no phase bias) at the B site for crystals soaked in 4-PBA at the indicated concentrations. No electron density is observable for ordered water or solute molecules at this contour level in the 0 mM 4-PBA experiment (F). Note how 4-PBA (colored purple) mimics the terminal phenylalanine side chain and carboxylate group in (E). To aid comparisons, the electron density calculations are all truncated at 2.8 Å; see Table 2 for details of X-ray datasets. See also Figure 1—figure supplement 1, and Tables 1 and 2.
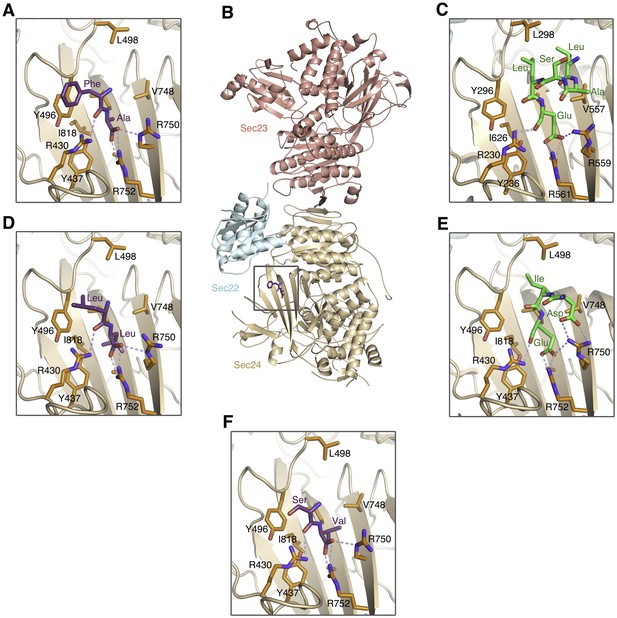
Comparison of the COPII binding site occupied by ΦC, DxE and LxxLE signal sequences.
(A) Close-up view of a bound ΦC sequence containing a terminal Phe-Ala motif (colored purple). This signal accelerates ER export of a reporter protein with potency comparable to Phe-Phe, according to Nufer et al. (2002). See Table 1 for details of peptide-bound crystal structures. (B) Structure of the COPII complex comprising human Sec23a/Sec24a•Sec22b bound to a ΦC peptide (boxed). The concave, membrane-binding surface of the coat faces the reader. (C) Picture of the equivalent binding site of S. cerevisiae Sec24 bound to the LxxLE motif of the SNARE protein Bet1 (colored green); from Mossessova et al. (2003). (D) A bound ΦC sequence containing a terminal Leu-Leu motif. Leu-Leu is an effective ER export motif, but not as potent as Val-Val (Nakamura et al., 1998; Nufer et al., 2002). (E) Picture shows the binding mode of the DxE motif of vesicular stomatitis virus G protein complexed with Sec24a (Mancias and Goldberg, 2008). (F) A bound ΦC sequence containing a terminal Ser-Val motif.
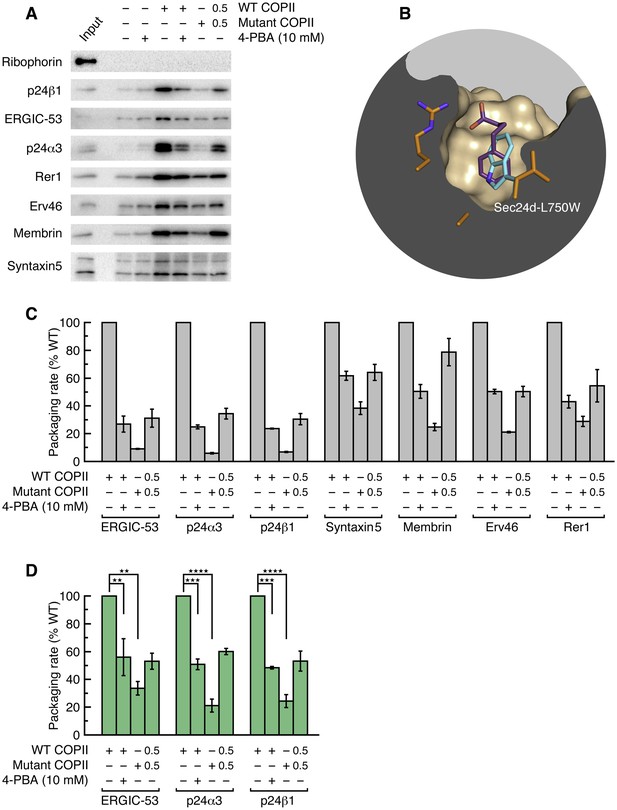
Application of 4-PBA or mutation of the B site of COPII reduces the packaging of p24 proteins in vesicles.
(A) Reconstituted COPII budding reactions were performed using permeabilized CHO (T-Rex-CHO-K1) cells, using either wild-type or mutant (Sec24d-L750W) COPII protein. Protein contents of the isolated vesicles were analyzed by immunoblotting. Lane labeled Input represents 1.1% of the starting permeabilized cell membranes used in a budding reaction; all other lanes represent the vesicle product of 100% of the starting membranes. The syntaxin5 antibody recognizes both the short and long forms of the SNARE protein. These are representative blots from experiments performed in triplicate. (B) Close-up view of the B site of Sec24d, oriented as in Figure 1E–I, showing 4-PBA (colored purple) and a modeled position of the tryptophan side chain (cyan) in the Sec24d-L750W mutant protein used in budding reactions. (C) The packaging rate for cargo proteins was determined by immunoblotting as in (A) and densitometry of chemiluminescent signals. For each cargo molecule, data are normalized to be a percentage of the packaging rate measured using wild-type COPII protein. Bar graphs show mean ± s.e.m. (n = 3). (D) Data in (C) were corrected for treatment-dependent (4-PBA or mutant COPII) changes in budding yield (using the mean values of the packaging rates for Syntaxin 5, membrin, Erv46 and Rer1). For presentation clarity, data are normalized as in (C). Statistical analysis was performed on data prior to normalization in order to preserve statistical distributions (n = 3; ANOVA test, Bonferroni-Holm post hoc, **p<0.01, ***p<0.001 and ****p<0.0001). Bar graphs show mean ± s.e.m. See also Table 3.
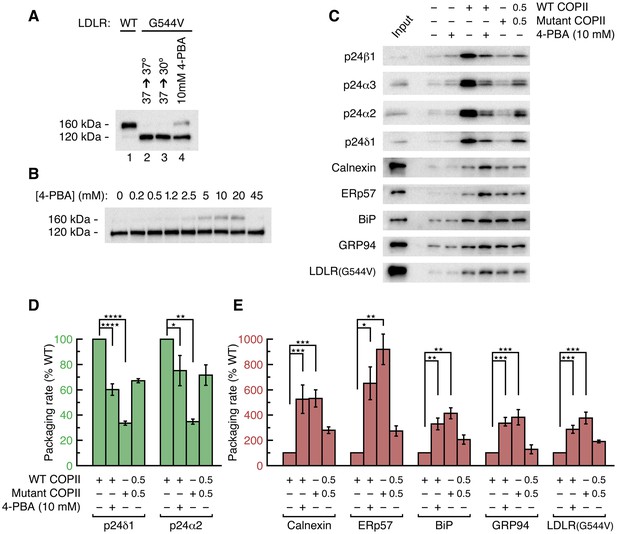
4-PBA promotes packaging of ER resident proteins and mutant LDL receptor into COPII vesicles in vitro.
(A) The G544V-mutant LDL receptor fails to reach the Golgi complex (Tveten et al., 2007). CHO cells stably transfected with wild-type (lane 1) or mutant LDL receptor were induced with tetracycline for 24 hr at 37°C before further incubation for 2 hr at 37°C (lane 2) or 30°C (lane 3), or 2 hr incubation at 37°C with 10 mM 4-PBA (lane 4). Cell lysates were analyzed by immunoblotting; labeling indicates the 160 kDa mature glycosylated and 120 kDa precursor forms of LDL receptor. (B) Dose-dependent restoration of trafficking of mutant LDL receptor by 4-PBA (see also Tveten et al., 2007). CHO cells expressing mutant LDL receptor were incubated with indicated concentrations of 4-PBA for 2 hr at 37°C. Cell lysates were analyzed as in (A). (C) The packaging of a series of p24 proteins, ER residents and mutant LDL receptor were assayed in scaled-up COPII budding reactions. Lane labeled Input represents 1.1% of the starting membranes; all other lanes represent the vesicle product of 100% of the starting membranes. Experiments were performed in triplicate. (D) Packaging rates for the p24-family proteins p24δ1 and p24α2, presented as in 2D (controlled by Syntaxin 5, membrin, Erv46 and Rer1 packaging rates). The data for p24α3 and p24β1 are in Figure 2D (n = 3; ANOVA test, Bonferroni-Holm post hoc, *p<0.05, **p<0.01 and ****p<0.0001). Bar graphs show mean ± s.e.m. (E) Graph shows packaging rates for four ER resident proteins and mutant LDL receptor. Note the change of scale of the y-axis (n = 3; ANOVA test, Bonferroni-Holm post hoc, *p<0.05, **p<0.01 and ***p<0.001). Bar graphs show mean ± s.e.m.
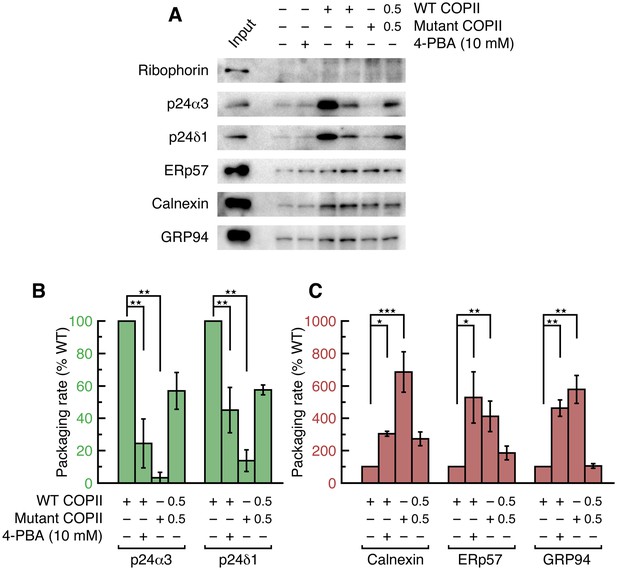
4-PBA causes packaging of ER resident proteins into COPII vesicles budded from unstressed cell membranes.
(A) Reconstituted COPII budding reactions were performed on permeabilized CHO cells (expressing neither wild-type nor mutant LDL receptor). Lane labeled Input represents 1.1% of the starting membranes used in a budding reaction; all other lanes represent the vesicle product of 100% of the starting membranes. Experiments were performed in triplicate. (B) Data in (A) were corrected as in Figure 2D, using the mean values of the packaging rates for Syntaxin 5, Erv46 and Rer1 (n = 3; ANOVA test, Bonferroni-Holm post hoc, **p<0.01). Bar graphs show mean ± s.e.m. (C) Graph shows packaging rates for Calnexin, ERp57 and GRP94 (n = 3; ANOVA test, Bonferroni-Holm post hoc, *p<0.05, **p<0.01 and ***p<0.001). Bar graphs show mean ± s.e.m.
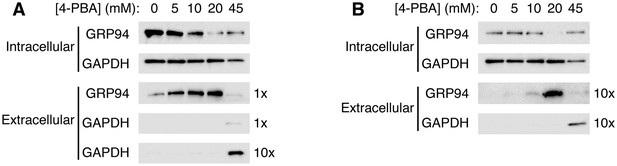
4-PBA causes the secretion of the KDEL-tagged luminal resident GRP94.
(A) CHO cells expressing G544V-mutant LDL receptor were incubated with increasing concentrations of 4-PBA for 24 hr at 37°C. Extracellular GRP94 in the medium was analyzed directly. Note that ER trapping of the mutant LDL receptor causes UPR leading to elevated levels of intracellular GRP94 (0 mM 4-PBA lane), and that UPR is attenuated at 10–20 mM 4-PBA (Sørensen et al., 2006). (B) Experiment used unstressed wild-type CHO cells. The blots in (A) and (B) showing intracellular levels of GRP94 are from equivalent amounts of cells (based on total protein measurement). To detect extracellular GRP94, ten times more sample was loaded than in (A).
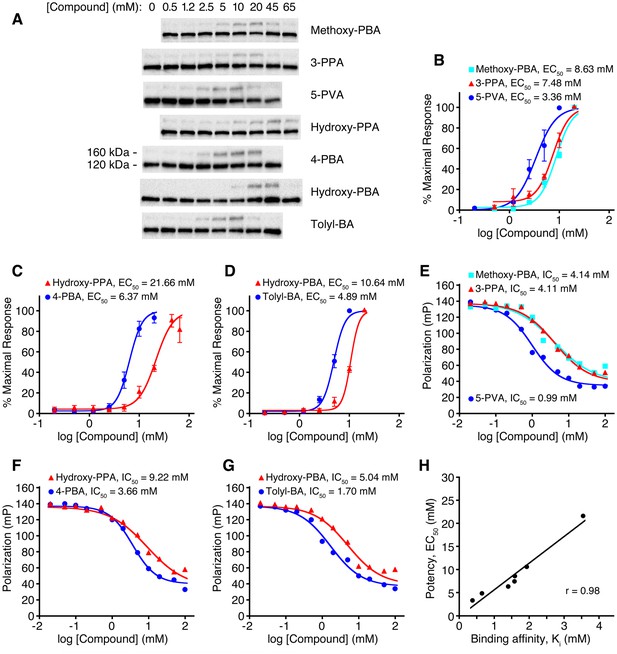
Relation between COPII binding-site affinity and pharmacological potency for a series of 4-PBA analogs.
(A) CHO cells expressing G544V-mutant LDL receptor were incubated with the indicated concentrations of 4-PBA analogs for 2 hr at 37°C. Cell lysates were analyzed by immunoblotting for the appearance of 160 kDa mature glycosylated LDL receptor (labeling indicates the position of 160 kDa mature glycosylated and 120 kDa precursor forms of LDL receptor). A gamma correction has been applied to the picture to improve clarity. (B) The acquisition of mutant LDL receptor glycosylation as a function of compound concentration is plotted and EC50 values reported. Data were obtained from (A) by densitometry of chemiluminescent signals. Methoxy-PBA is 4-(4-methoxyphenyl)butyrate (n = 4 independent experiments); 3-PPA, 3-phenylpropionate (n = 4); 5-PVA, 5-phenylvalerate (n = 5). Standard errors for EC50 values are reported in Materials and methods. (C) Hydroxy-PPA, 3-(4-hydroxyphenyl)propionate (n = 4); 4-PBA (n = 5). (D) Hydroxy-PBA, 4-(4-hydroxyphenyl)butyrate (n = 4); Tolyl-BA, 4-(4-tolyl)butyrate (n = 4). (E–G) Displacement of the fluorescent probe 5-Fam-QIYTDIEANR (based on the ER export signal of VSV G protein) from human Sec24a by 4-PBA analogs was measured by fluorescence polarization at 22°C. IC50 values were determined by non-linear regression fitting (standard errors are reported in Materials and methods). (H) Scatter plot compares data from the cell-based and fluorescence polarization experiments. For the latter, IC50 values were converted to true affinities, Ki, using the approach of Nikolovska-Coleska et al., 2004. See also Figure 6—figure supplement 1.
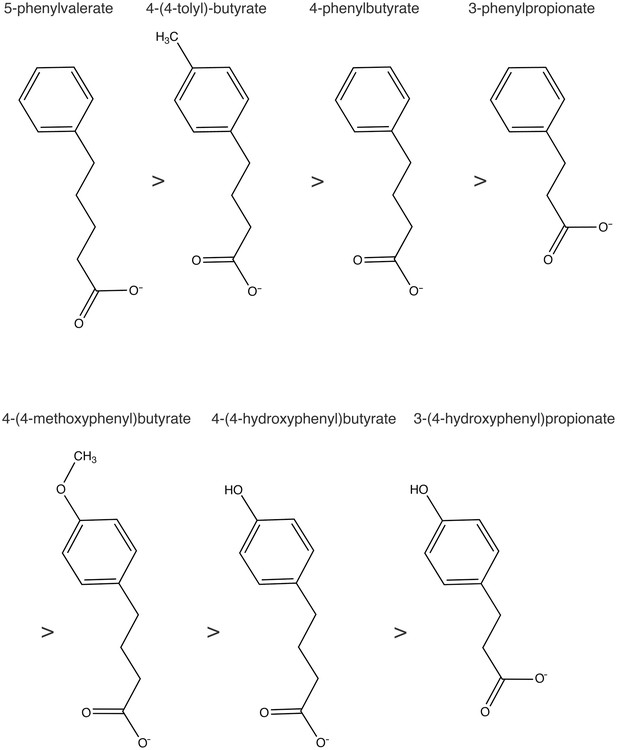
The series of 4-PBA analogs.
The structural formulae for 4-PBA analogs are drawn in order of their affinity for COPII protein.
Tables
Data Collection and Refinement Statistics – ΦC signal complexes.
| COPII protein complex | Sec23/24•22 | Sec23/24•22 | Sec23/24•22 | Sec23/24•22 | Sec23/24•22 | Sec23/24•22 | Sec23/24•22 |
| Peptide | EVTSLV | EVTSVV | EVTSII | EVTSSV | EVTSFA | EVTSFF | EVTSLL |
| Protein Data Bank ID | 5VNE | 5VNF | 5VNG | 5VNH | 5VNI | 5VNJ | 5VNK |
| Space group: | C2 | C2 | C2 | C2 | C2 | C2 | C2 |
| copies/asymmetric unit | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cell parameters a, b, c (Å) | 148.4, 96.8, 127.0 | 147.8, 97.1, 128.8 | 148.0, 96.9, 130.0 | 147.9, 96.9, 127.3 | 147.6, 96.1, 130.0 | 147.9, 96.9, 129.5 | 148.2, 97.6, 130.5 |
| Cell parameters β (°) | 91.7 | 90.1 | 90.1 | 91.6 | 90.1 | 90.2 | 90.1 |
| Data processing | |||||||
| Resolution (Å) | 50–2.7 | 50–2.6 | 50–2.6 | 50–2.6 | 50–2.8 | 50–2.8 | 50–2.6 |
| Rmerge (%)† | 8.9 (70.2) | 9.6 (52.0) | 11.4 (55.0) | 8.0 (77.4) | 11.6 (58.2) | 11.6 (58.2) | 12.6 (78.9) |
| I/σ | 24.5 (2.6) | 18.0 (1.5) | 16.2 (1.1) | 29.5 (2.5) | 12.6 (1.0) | 12.6 (1.0) | 30.1 (2.3) |
| Completeness (%) | 99.8 (100) | 99.0 (99.5) | 98.8 (99.2) | 99.5 (100) | 98.2 (99.5) | 98.2 (99.5) | 99.5 (99.0) |
| Redundancy | 4.0 (4.0) | 3.7 (3.1) | 3.2 (2.4) | 4.2 (4.2) | 2.5 (2.3) | 2.5 (2.3) | 3.8 (3.7) |
| Refinement statistics | |||||||
| Data range (Å) | 50–2.7 | 50–2.6 | 50–2.6 | 50–2.6 | 50–2.8 | 50–2.8 | 50–2.6 |
| Reflections | 52341 | 54635 | 54087 | 61690 | 40031 | 44706 | 60273 |
| Nonhydrogen atoms | 12597 | 12619 | 12558 | 12577 | 12545 | 12535 | 12559 |
| Water molecules | 99 | 111 | 59 | 81 | 46 | 77 | 60 |
| R.m.s. ∆ bonds (Å)‡ | 0.6 | 0.6 | 0.7 | 0.6 | 0.6 | 0.5 | 0.6 |
| R.m.s. ∆ angles (°)‡ | 0.002 | 0.003 | 0.002 | 0.002 | 0.003 | 0.002 | 0.002 |
| R-factor (%)§ | 22.2 | 20.5 | 20.7 | 21.5 | 21.7 | 21.3 | 19.6 |
| Rfree (%)§, # | 26.8 | 24.1 | 25.7 | 25.3 | 27.3 | 26.0 | 25.1 |
-
*Highest resolution shell is shown in parenthesis.
-
†Rmerge = 100 x ∑h∑i | Ii(h) - <I(h)> | / ∑h<I(h)>, where Ii(h) is the ith measurement and <I(h)> is the weighted mean of all measurement of I(h) for Miller indices h.
-
‡Root-mean-squared deviation (r.m.s. ∆) from target geometries.
-
§R-factor = 100 x ∑|FP – FP(calc)|/∑ FP.
-
#Rfree was calculated with 5% of the data.
Data collection and refinement statistics – 4-PBA crystallography.
| COPII protein complex | Sec23/24•22 | Sec23/24•22 | Sec23/24•22 | Sec23/24•22 |
| 4-PBA Concentration | 1 mM | 15 mM | 50 mM | 0 mM |
| Protein Data Bank ID | 5VNL | 5VNM | 5VNN | 5VNO |
| Space group | C2 | C2 | C2 | C2 |
| Copies/asymmetric unit | 1 | 1 | 1 | 1 |
| Cell parameters: a, b, c (Å) | 147.8, 96.8, 129.6 | 149.1, 96.8, 130.6 | 148.02, 97.7, 128.9 | 148.4, 96.9, 130.5 |
| cell parameters: β (°) | 90.2 | 90.2 | 89.6 | 89.6 |
| Data processing | ||||
| Resolution (Å) | 50–2.4 | 50–2.8 | 50–2.5 | 50–2.9 |
| Rmerge (%)† | 8.9 (45.9) | 9.9 (80.9) | 9.3 (56.7) | 7.6 (86.2) |
| I/σ | 16.1 (1.2) | 11.6 (1.2) | 13.3 (1.3) | 15.9 (1.7) |
| Completeness (%) | 98.9 (95.9) | 99.3 (99.5) | 99.2 (99.1) | 98.6 (99.5) |
| Redundancy | 2.9 (1.8) | 3.7 (3.9) | 3.0 (2.3) | 3.6 (3.8) |
| Refinement statistics | ||||
| Data range (Å) | 50–2.4 | 50–2.8 | 50–2.5 | 50–2.9 |
| Reflections | 61011 | 43047 | 53119 | 39048 |
| Nonhydrogen atoms | 12471 | 12462 | 12491 | 12477 |
| Water molecules | 50 | 45 | 59 | 48 |
| R.m.s. ∆ bonds (Å)‡ | 0.6 | 0.6 | 0.6 | 0.6 |
| R.m.s. ∆ angles (°)‡ | 0.003 | 0.002 | 0.002 | 0.003 |
| R-factor (%)§ | 21.1 | 21.3 | 21.2 | 20.9 |
| Rfree (%)§, # | 24.6 | 26.0 | 25.1 | 25.3 |
-
*Highest resolution shell is shown in parenthesis.
-
†Rmerge = 100 x ∑h∑i | Ii(h) - <I(h)> | / ∑h<I(h)>, where Ii(h) is the ith measurement and <I(h)> is the weighted mean of all measurement of I(h) for Miller indices h.
-
‡Root-mean-squared deviation (r.m.s. ∆) from target geometries.
-
§R-factor = 100 x ∑|FP – FP(calc)|/∑ FP.
-
#Rfree was calculated with 5% of the data.
Cargo packaging efficiency measured for wild-type COPII protein.
| Packaging efficiency (Amount in vesicle as % of protein present in starting permeabilized cells) | ||
|---|---|---|
| CHO-LDLRG544V cells | Wild-Type CHO cells | |
| Cargo | ||
| ERGIC-53 | 4.5 | – |
| p24δ1 | 7.7 | 4.6 |
| p24α2 | 8.7 | 10.9 |
| p24β1 | 10.6 | – |
| p24α3 | 6.4 | 6.4 |
| Syntaxin 5 | 0.89 | 0.96 |
| Membrin | 4.8 | – |
| Erv46 | 6.6 | 3.9 |
| Rer1 | 11.5 | 5.4 |
| ER resident | ||
|---|---|---|
| Calnexin | 0.32 | 0.065 |
| ERp57 | 0.63 | 0.12 |
| BiP | 0.87 | – |
| GRP94 | 0.25 | 0.07 |





