Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80
Figures
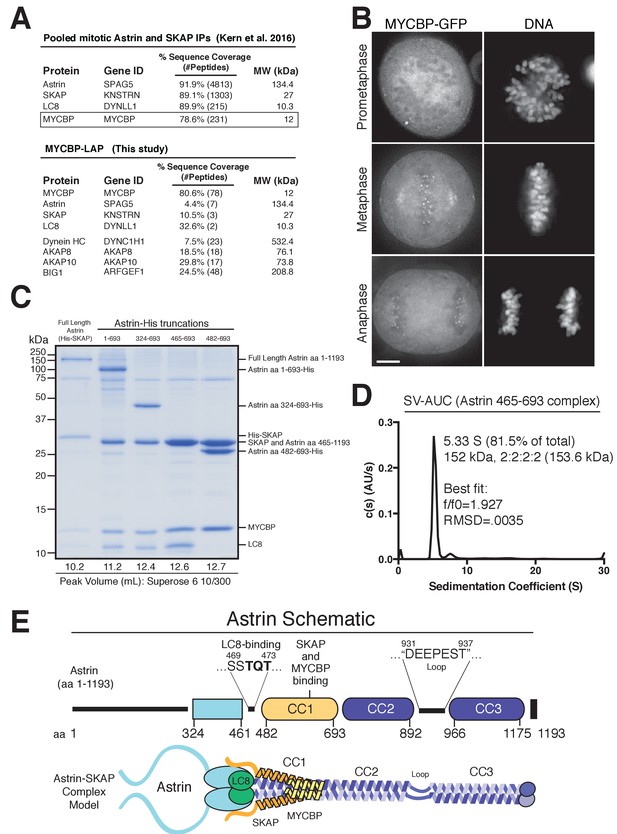
Reconstitution of a 4-subunit Astrin-SKAP complex.
(A) Top: Pooled mass spectrometry data from Astrin and SKAP IPs (Kern et al., 2016) identifies MYCBP. Bottom: MYCBP IP (this study) isolates Astrin-SKAP complex components. (B) MYCBP-GFP localizes to the mitotic spindle and aligned kinetochores. Deconvolved image sections of selected MYCBP-GFP cells in mitosis were selected and scaled individually to show localization. Many cells from multiple experiments were analyzed with live and fixed cell microscopy to draw conclusions about MYCBP localization. Also see Figure 1—figure supplement 1A. Scale bar, 5 µm. (C) Coomassie gel of Astrin-SKAP complex purifications. Complex components and truncations were co-expressed using the MultiBac system in SF9 cells. Indicated His-tags were used for complex purification followed by gel filtration. For each complex, gel filtration peaks were pooled and spin concentrated before polyacrylamide gel loading. Gel filtration peak migration volumes are given below each lane (also see Figure 1—figure supplement 1B). Void Volume: 8.5 mL, Thyroglobulin (8.5 nm stokes radius) Size Standard: 13.1 mL. (D) Sedimentation velocity ultracentrifugation of the Astrin 465–693 complex. The complex was fit to a single major peak with the indicated statistics. (E) Schematic of Astrin-SKAP complex structure based on data in this figure and Figure 1—figure supplement 1.
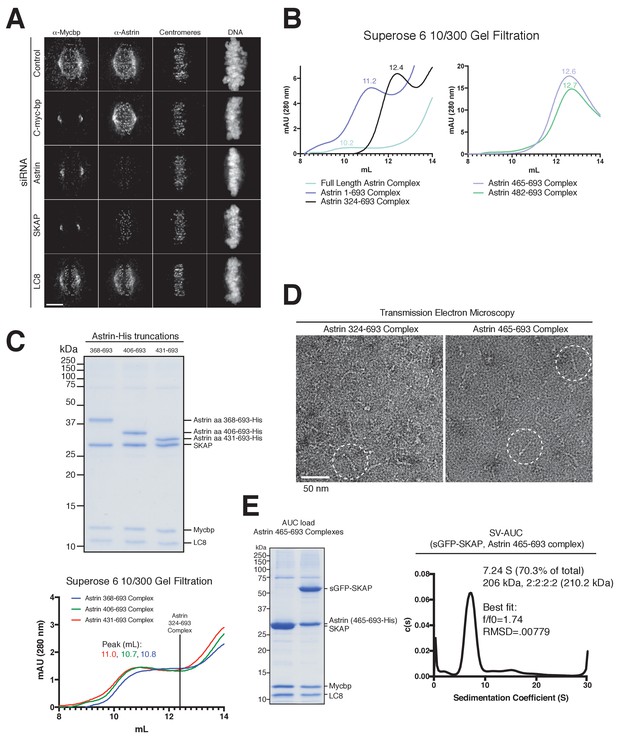
MYCBP kinetochore localization is dependent on Astrin and SKAP and Astrin-SKAP complex biochemical characterization.
(A) Localization dependencies of MYCBP in the Astrin-SKAP complex. Note: In conditions where substantial phenotypic defects were present (Astrin and SKAP depletion), rare cells with metaphase alignment were analyzed to control for the effect of alignment on Astrin-SKAP complex localization. Immunofluorescence images are deconvolved projections through the mitotic spindle. Individual channel levels are scaled equivalently across siRNA experiments. Fixation (PEPF). Scale bar, 5 µM. (B) Superose six gel filtration traces for the indicated Astrin-SKAP complexes. Peak migration (mL) is given above each peak. Void Volume: 8.5 mL, Thyroglobulin (8.5 nm stokes radius) Size Standard: 13.1 mL. (C) Top: Coomassie gel of Astrin-SKAP complex purifications as in Figure 1C. Bottom: Superose six gel filtration traces as in part B. The volume where the Astrin 324–693 complex migrates is indicated with a line for comparison. (D) Negative stain transmission electron microscopy for the Astrin 324–693 and Astrin 465–693 complexes. (E) Left: Coomassie gel of the Astrin 465–693 complexes loaded for AUC analysis. Right: Sedimentation velocity ultracentrifugation of the Astrin 465–693 complex with sfGFP-SKAP. The complex was fit to a single major peak with indicated statistics.
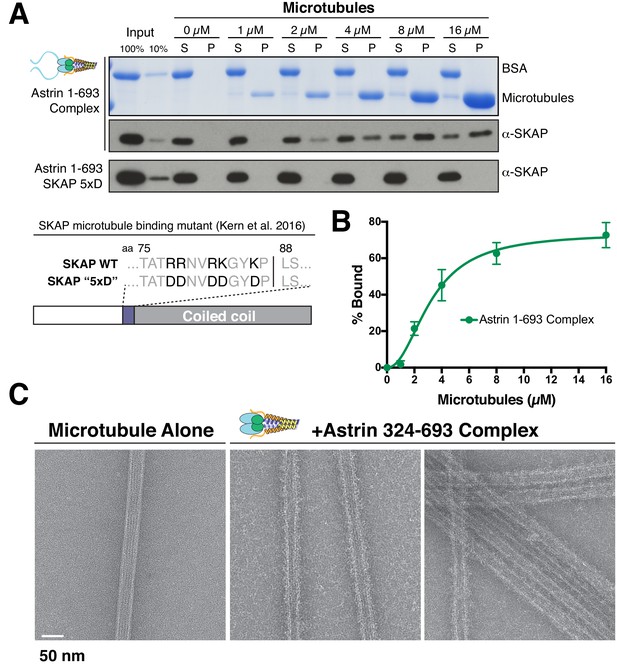
The Astrin-SKAP complex binds to microtubules through its SKAP microtubule-binding domain.
(A) Top: Astrin 1–693 complex microtubule binding assays showing the Coomassie stained gel and α-SKAP Western blots from an example experiment. Bottom: Equivalent assay for the SKAP 5xD mutant version of the complex with the α-SKAP Western blot shown. Mutated residues for the 5xD mutant are illustrated below (also see [Kern et al., 2016]). (B) Microtubule-binding curve generated from triplicate binding experiments. Mean and standard deviation are plotted from quantified Western blots (see Materials and methods). The data was fit in Prism using the model of specific binding with a Hill slope. (C) Images of Astrin-SKAP complex-bound microtubules visualized with negative-stain electron microscopy.
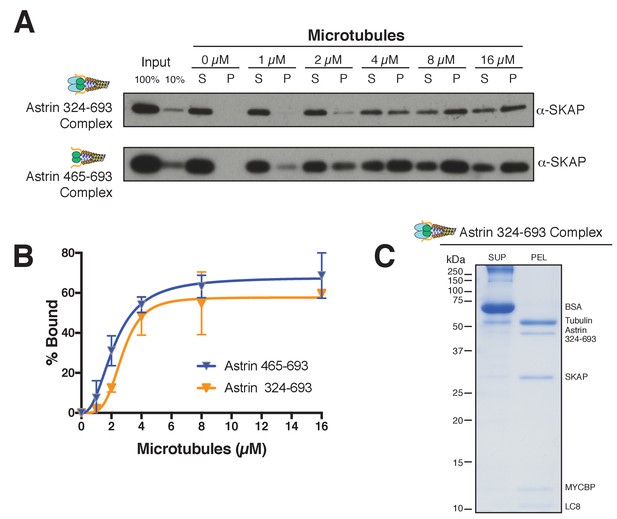
Astrin truncations that associate with SKAP are sufficient to bind microtubules in vitro.
(A) Examples of Astrin-SKAP complex microtubule binding assays showing α-SKAP Western blots for the indicated constructs. (B) Microtubule-binding curves generated from triplicate (Astrin 465–693) or duplicate (Astrin 324–693) binding experiments as shown in part A. Mean and standard deviation are plotted from quantified Western blots (see Materials and methods). The data was fit in Prism using the model of specific binding with a Hill slope. (C) High concentration, Coomassie-based microtubule binding assay for the Astrin 324–693 complex. Microtubules, 2 µM; Astrin 324–693 complex,~0.5 µM.
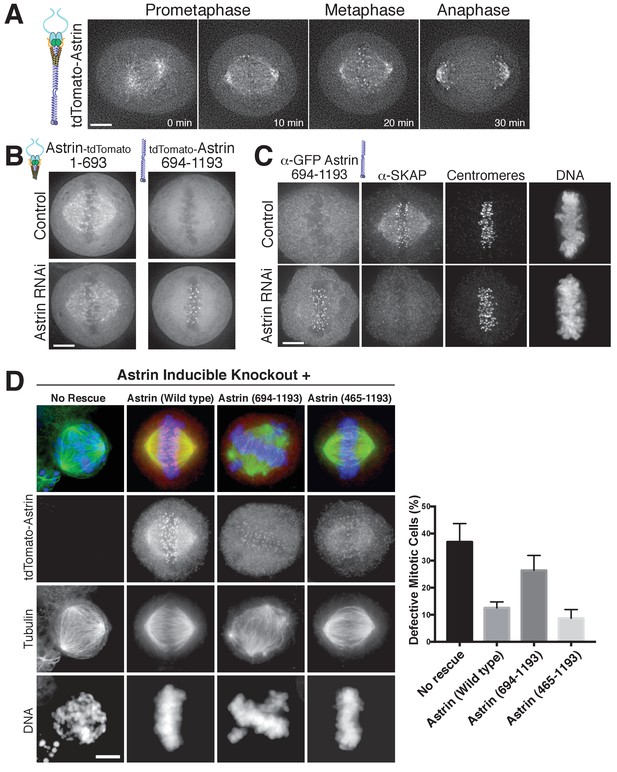
The Astrin-SKAP complex docks at kinetochores using two distinct domains.
(A) Time lapse images showing localization of tdTomato-Astrin, introduced with the BacMam system into HeLa cells, during a mitotic time course (see Figure 3—video 1). Images represent deconvolved sections. (B) Localization of the indicated Astrin constructs, introduced with the BacMam system, in control or Astrin-depleted cells. Images are deconvolved sections and are scaled equivalently for each construct. (C) Immunofluorescence images for control or Astrin-depleted cells expressing GFP-Astrin 694–1193 with the indicated antibodies. Each image is a deconvolved and projected image stack through the mitotic spindle. Cells were scaled equivalently for each antibody, and DNA (Hoechst-stain) was scaled individually. Fixation (PF). (D) Astrin inducible knockouts with BacMam replacements. Left: deconvolved and projected immunofluorescence images of representative phenotypes for the indicated conditions. Images were scaled equivalently for Astrin antibody and individually for DNA (Hoechst-stain) and microtubules. Right: Quantification of defective mitotic cells from two experiments per condition (200 cells counted per experiment) showing the percent of cells with mis-aligned chromosomes and/or multi-polar spindles. Scale bars, 5 µm.
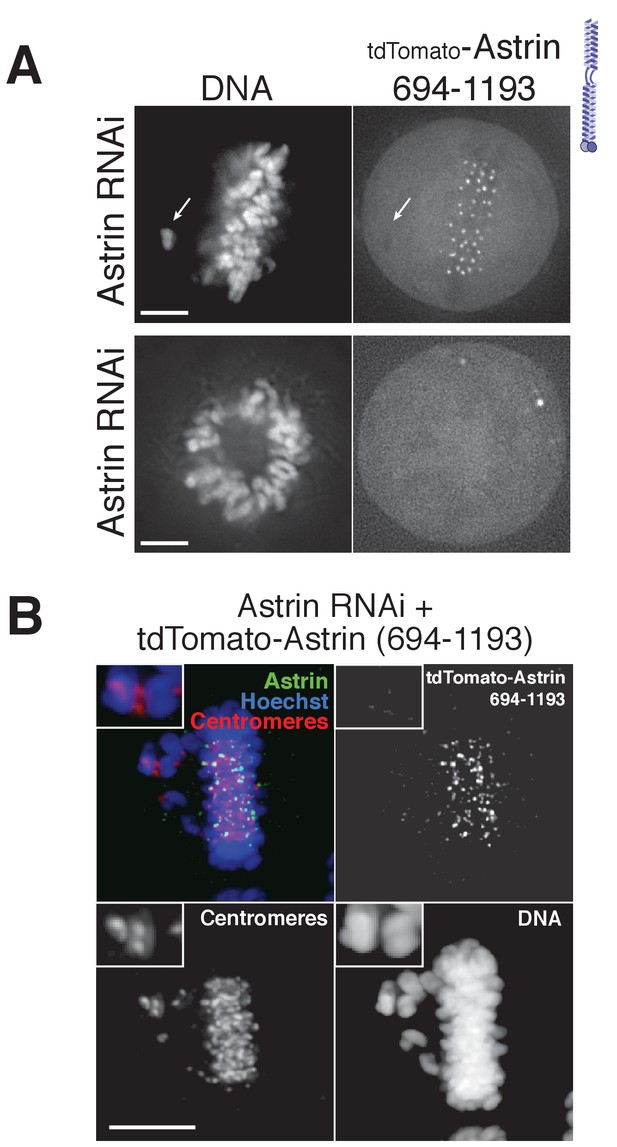
Astrin C-terminal domain (amino acids 694–1193) localizes to aligned kinetochores.
(A) Live cell imaging. Top: An off-axis chromosome is indicated, where there is no clear kinetochore localization compared to the aligned chromosomes in the same image. This image is a deconvolved stack projected through the misaligned chromosome. Bottom: The Astrin C-terminus (694–1193) does not localize to kinetochores in a prometaphase cell. Image is a single deconvolved section. Astrin 694–1193 was introduced with BacMam transduction. (B) Fixed-cell immunofluorescence showing lack of Astrin 694–1193 on mis-aligned kinetochores in an Astrin-depleted cell with anti-centromere antibody (ACA) as a kinetochore marker. Astrin 694–1193 was introduced using lentivirus (see Materials and methods), fixed in the same manner as cells in Figure 3D. These cells were subjected to a thymidine block 24 hr after siRNA addition and imaged at 48 hr using a Nikon 60x oil objective. Scale bars, 5 µm.
Astrin localizes to aligned kinetochores during mitosis.
Time lapse movie showing tdTomato-Astrin in red and DNA (Hoescht) in blue. See Figure 3A and Materials and methods.
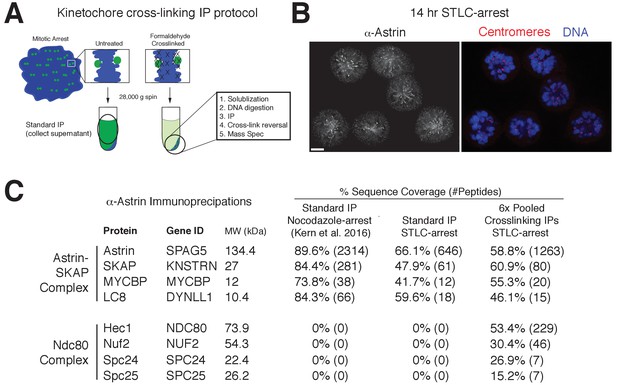
Kinetochore cross-linking mass spectrometry identifies Ndc80 as an Astrin-SKAP complex interaction partner.
(A) Schematic of kinetochore cross-linking IP protocol (see Materials and methods). (B) Immunofluorescence of STLC-arrested HeLa cells showing Astrin localization to kinetochores using the α-Astrin antibody utilized for IPs. Fixation (M). Scale bar, 5 µm. (C) α-Astrin immunoprecipitation-mass spectrometry with the indicated arrests. The cross-linking IPs (right), shown as combined data from six independent experiments, reveal an interaction between the Astrin-SKAP and Ndc80 complexes. See Figure 4—figure supplement 1 for additional data from the individual preparations and the Source dataset-Mass Spectrometry Data Supplement for the complete search information.
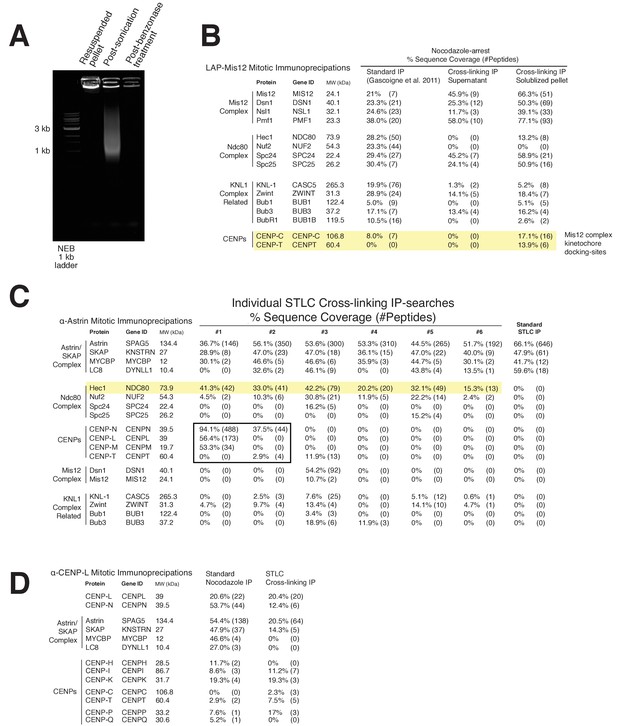
Kinetochore cross-linking mass spectrometry reveals kinetochore binding sites for dynamic interactions.
(A) Ethidium bromide-stained agarose gel showing the progression of DNA-destruction from the initial lysed-cell pellet for the crosslinking protocol. (B) LAP-Mis12 immunoprecipitations showing a comparison of the data of a standard preparation to the supernatant of a cross-linking preparation and the final kinetochore cross-linking protocol. (C) Mass spectrometry data from individual α-Astrin crosslinking IPs compared to the standard STLC prep (Pooled data is shown in Figure 4). (D) Mass spectrometry data from α-CENP-L cross-linking and standard IPs. Note: We analyzed and selected specific protein interactors from our mass spectrometry searches for presentation in this figure, taking into account common contaminants in similar unrelated samples and acceptable minimum sequence coverage from multiple kinetochore immunoprecipitations. Also see the provided full mass spectrometry search results.
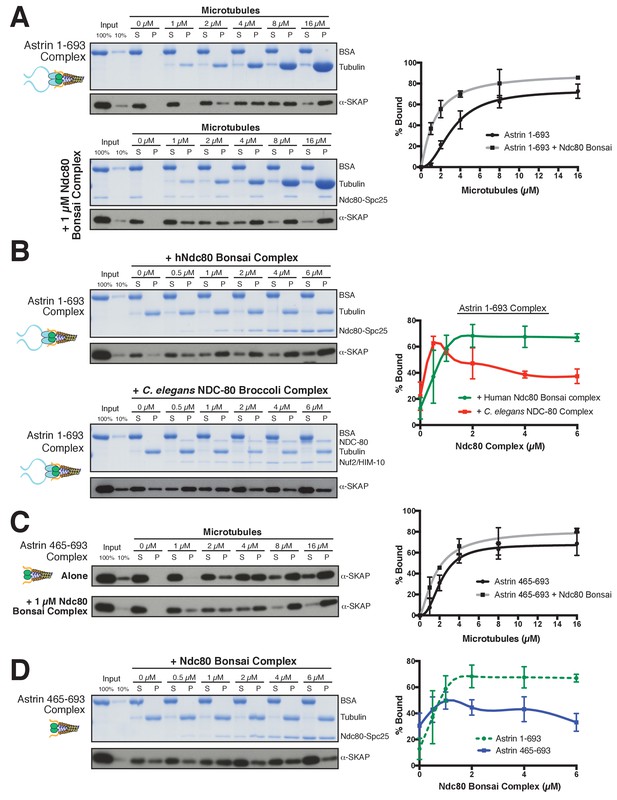
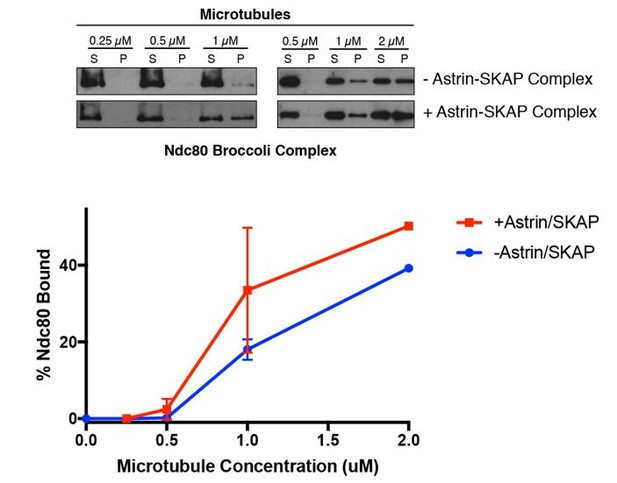
The Astrin-SKAP complex interacts with the Ndc80 complex in the presence of microtubules.
(A) Left: Example of Coomassie-stained gel and western blot (the same gel was cut to generate each for loading comparison) for the Astrin 1–693 complex bound to microtubules in the absence (top) and presence of 1 µM Ndc80 Bonsai complex. Right: Quantification of Astrin-SKAP microtubule binding from triplicate experiments. Curve for microtubule binding without Ndc80 was reproduced from Figure 2B. Mean and standard deviation are plotted, and the data was fit with Prism using the model of specific binding with a Hill slope (B) Left: Ndc80 complex saturation experiment example for the Astrin 1–693 complex with human Ndc80 Bonsai complex (top) and the C. elegans Ndc80 Broccoli complex (bottom). Microtubules, 2 µM. Right: Quantification of binding from triplicate (human) or duplicate (C. elegans) experiments. Mean and standard deviation are plotted, and the trend line was generated from a coarse LOWESS fit in prism. (C) Left: Western blots of microtubule binding (as in part A) with the Astrin 465–693 complex. Right: Quantification as in part A, Curve for microtubule binding without Ndc80 was reproduced from Figure 2—figure supplement 1B. (D) Left: Ndc80 Bonsai complex saturation experiment for the Astrin 465–693 complex. Microtubules, 2 µM. Right: Quantification of binding from triplicate experiments as in (B). The curve from part B for the Astrin 1–693 complex is duplicated as a dotted line for comparison.
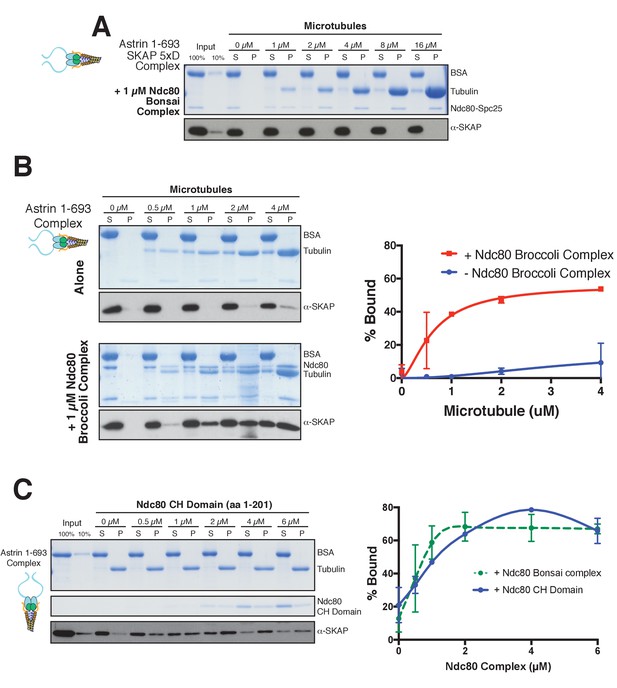
Astrin-SKAP microtubule binding is necessary for its co-pelleting with the Ndc80-microtubule complex and the Astrin-SKAP complex interacts with the microtubule-binding Ndc80 CH-domain.
(A) Coomassie gel and Western blot showing the absence of microtubule binding in the presence of Ndc80 Bonsai for the 5xD SKAP mutant, Astrin 1–693 complex. (B) Left: Coomassie gel and Western Blot showing an Astrin 1–693 complex microtubule binding experiment with and without added Ndc80 Broccoli complex. Right: Quantification of binding from duplicate experiments. Mean and standard deviation are plotted from 2 independent experiments, and the trend line was generated from a coarse LOWESS fit in prism. (C) Top: Microtubule binding for the Astrin 1–693 complex in the presence of the Ndc80 CH-domain. Microtubules, 2 µM. Bottom: Quantification of Astrin-SKAP complex binding in the CH-domain binding experiment (mean and standard deviation from two independent experiments) compared to the Ndc80 Bonsai curve (reproduced from Figure 5C).
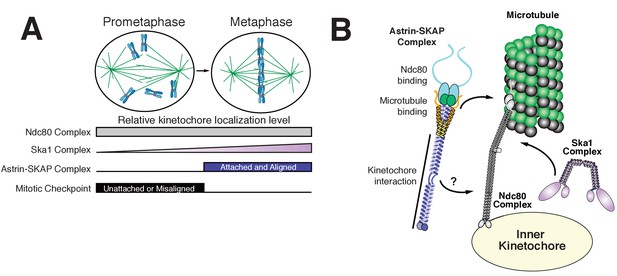
Model of Astrin-SKAP kinetochore attachment.
(A) Schematic of kinetochore localization in prophase and metaphase for a selection of outer kinetochore components. Ndc80 complex localizes during mitosis to all kinetochores and the Ska1 complex kinetochore localization increases as mitosis proceeds. The Astrin-SKAP complex and the proteins of the mitotic checkpoint each show switch-like kinetochore localization, but with opposite timing. Astrin-SKAP complex localizes strongly to microtubule-bound and bioriented kinetochores, whereas mitotic checkpoint proteins localize preferentially to unattached and misaligned kinetochores. (B) The Astrin-SKAP complex binds to kinetochores through SKAP microtubule binding, Astrin’s N-terminus binding to Ndc80 complex, and Astrin’s C-terminus binding to an unknown kinetochore location. Through these multiple interactions, the Astrin-SKAP complex can provide an intra-kinetochore tether to bridge the kinetochore and stabilize microtubule interactions. Together, the Ska1, Ndc80, and Astrin-SKAP complexes form the major microtubule binding interface of metaphase human kinetochores.
Additional files
-
Source data 1
Source dataset-Mass spectrometry data.
Complete mass spectrometry searches using methods described in (Washburn et al., 2001) for affinity purification/mass spectrometry data sets described in this paper (data from this study; [Kern et al., 2016] [Gascoigne et al., 2011]). Individual Astrin cross-linking immunoprecipitations are listed based on the order in Figure 4—figure supplement 1. These samples have not been pruned for common or antibody-specific contaminants.
- https://doi.org/10.7554/eLife.26866.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26866.015