Dual control of pcdh8l/PCNS expression and function in Xenopus laevis neural crest cells by adam13/33 via the transcription factors tfap2α and arid3a
Figures
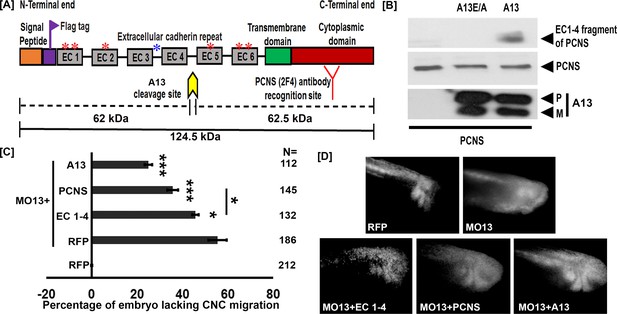
adam13 cleaves pcdh8l.
(A) Schematic representation of full length pcdh8l. A Flag tag (DYKDDDDK) was introduced just before the first cadherin repeat (EC1). The yellow arrow indicates the predicted cleavage site of adam13 based on the molecular weight of N- and C- terminal fragments. Red and blue asterisk indicate N and O glycosylation site, respectively. The monoclonal antibody 2F4 was produced against the cytoplasmic domain (Red). (B) Western blot from transfected Hek293T cells. Glycoproteins from the conditioned media were purified on concanavalin-A agarose. The cleavage fragment was detected using the N-terminus Flag tag with the mAb M2. Total pcdh8l and adam13 were detected using mAb 2F4 and mAb 4A7, respectively. The Pro (P) and Metalloprotease (M) active forms of adam13 are indicated. (C) Histogram representing the percentage of embryos lacking fluorescent neural crest cell in the migration pathway. (D) Photographs of representative embryos with or without fluorescent migrating neural crest cells. Embryos were injected at the one-cell stage with a morpholino targeting adam13 (10 ng MO13). Messenger RNA for RFP alone or combined with the different constructs was injected at the 8-cell-stage in a dorsal animal blastomere. Each injection was compared to RFP injected in control embryos in which RFP positive cranial neural crest cells have successfully migrated into the branchial and hyoid arches (0% inhibition). N = number of embryos scored from three or more independent experiments. Error bars represent standard error to the mean. One-way ANOVA was performed to determine statistical significance. Statistically significant at *p<0.05, ***p<0.005.
-
Figure 1—source data 1
Source data for Figure 1.
- https://doi.org/10.7554/eLife.26898.005
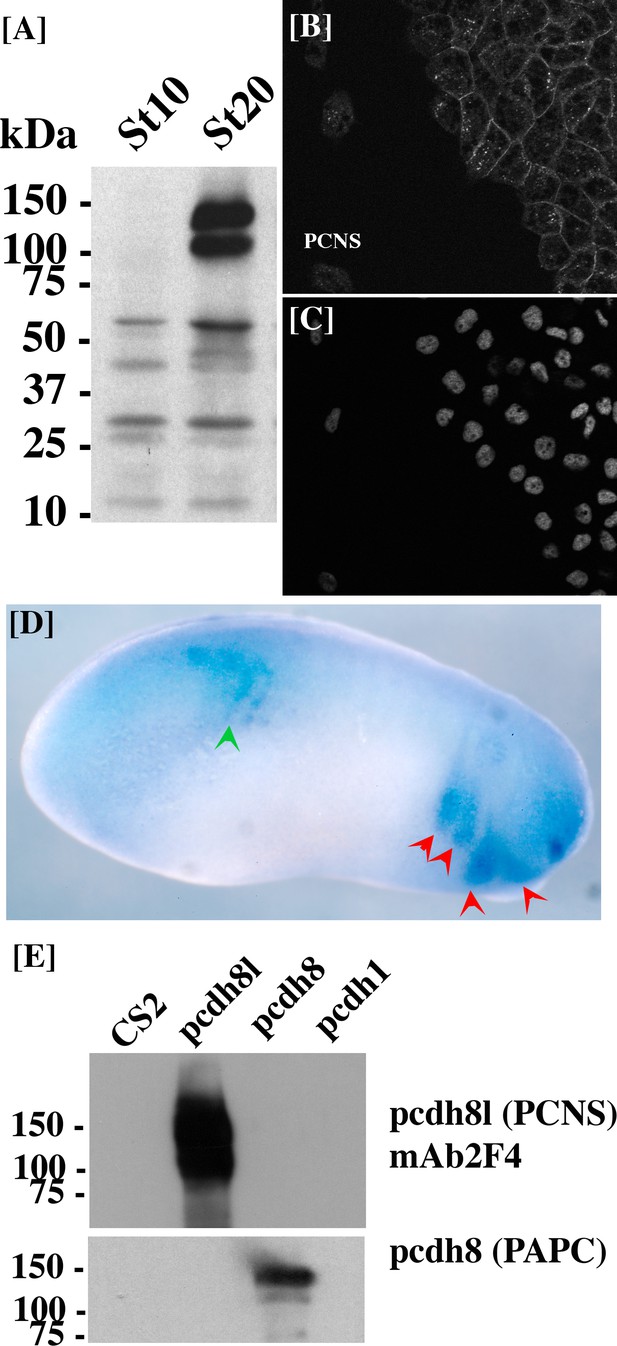
Characterization of mAb 2F4 to pcdh8l.
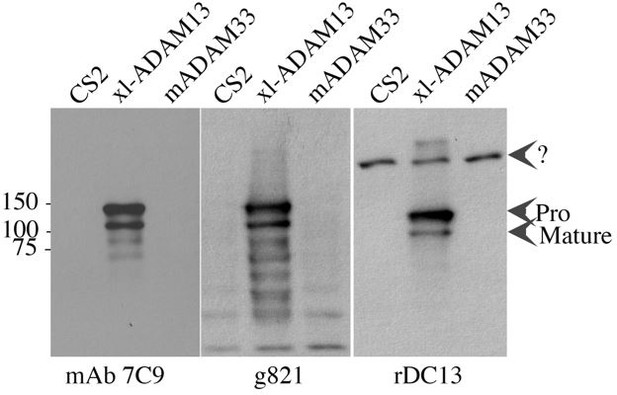
(A) Western blot from total membrane extracted from 10 embryos at gastrula stage (Stage 10) and early tailbud stage (Stage 20) detected using mAb2F4. (B–C) Cranial neural crest explants on fibronectin. Explants were left to migrate for 5 hr and fixed in 1XMBS, 3.7% formaldehyde for 1 hr, permeabilized for 30 min in 1XMBS containing 0.5% TritonX100. Staining with mAb 2F4 (B) clearly shows a membrane staining while DAPI stains the nuclei (C). (D) Wholemount immunostaining of tailbud stage embryo fixed in DENTS (20% DMSO 80% Methanol) using mAb 2F4. The red arrows indicate the tip of the cranial neural crest segments. The staining is identical to the one obtained by in situ hybridization with the pcdh8l probe. (E) Western blot on Hek293T cells extract transfected with the various Xenopus laevis protocadherin. The mAb 2F4 only recognizes pcdh8l (PCNS).
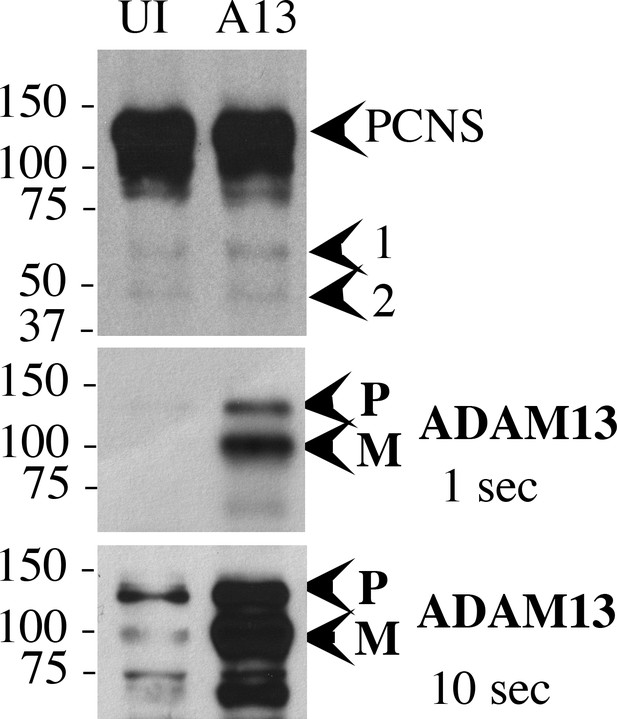
adam13 overexpression increases pcdh8l fragments.
Western blot with mAb 2F4 from glyctoproteins isolated from 20 embryos at stage 20. The intact full-length pcdh8l protein (PCNS) is indicated as well as two main fragments (arrowhead 1 and 2). All three forms are detected in uninjected embryos (UI) but the fragments are increased in embryos overexpressing adam13 (A13). The blot was re-probed with 15F, a polyclonal antibody directed against the adam13 cytoplasmic domain (Alfandari et al., 1997). The Pro (P) and Mature (M) forms of adam13 are indicated. Two exposures are provided to clearly detect the endogenous and overexpressed adam13 protein.
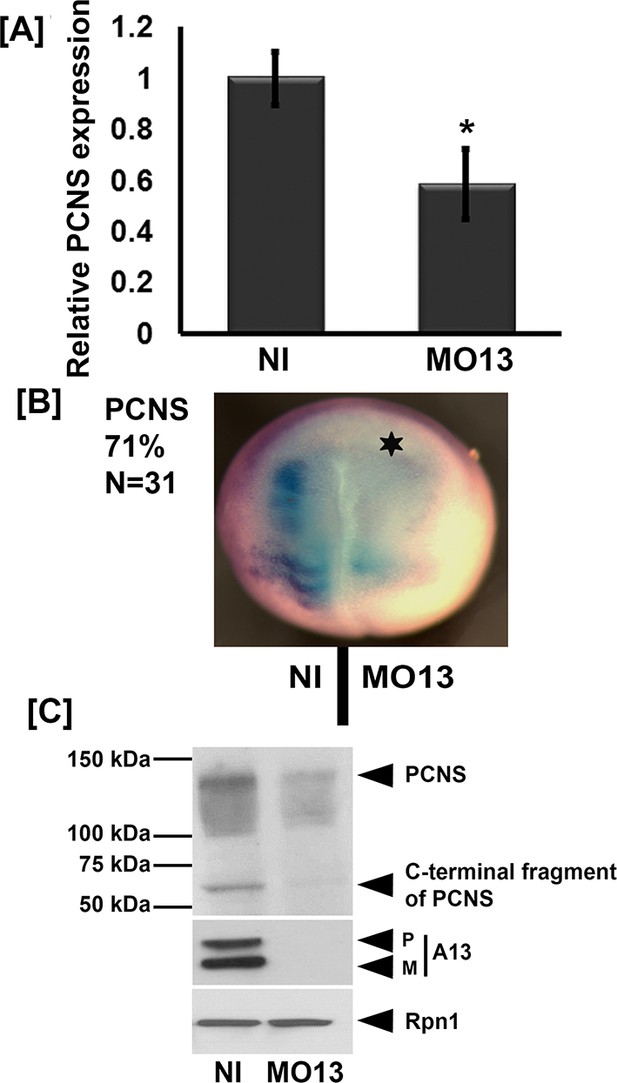
Adam13 knockdown reduces pcdh8l expression.
(A) Relative mRNA expression of pcdh8l normalized to GAPDH obtained by real-time PCR. MO13 was injected at the one cell stage (10 ng) and embryos were collected at stage 20. Polyadenylated mRNA was extracted from non-injected and MO13 injected embryos (five embryos each). (B) Representative images of whole mount in situ hybridization in which MO13 (5 ng) was injected into one of the two blastomeres and non-injected side serves as control. Image shows reduced mRNA levels of pcdh8l in MO13 injected side (star, 71% of 31 embryos). (C) Western blot on glycoproteins isolated from either control (Non Injected) or knock down (10 ng MO13) stage 20 embryos (25 embryos each). MO13 efficiently prevents the translation of adam13 (A13) as well as reduces the pcdh8l protein level. A monoclonal antibody to Xenopus Ribophorin 1 (Rpn1) was used as a loading control. Error bars represent standard error to the mean (Mean ±S.E.M). One-way ANOVA was performed to determine statistical significance. Statistically significant at *p<0.05.
-
Figure 2—source data 1
Source data for Figure 2.
- https://doi.org/10.7554/eLife.26898.009
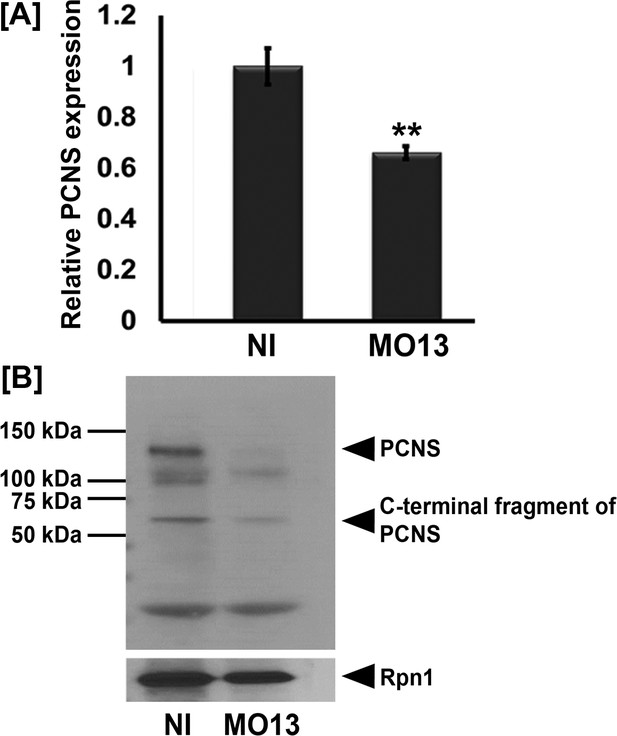
adam13 knockdown reduces pcdh8l expression in cranial neural crest (CNC) cells.
(A) Relative mRNA expression of pcdh8l normalized to GAPDH in isolated CNC explants. CNC explants (Alfandari et al., 2003) were dissected from either control embryos or embryos injected with MO13 (10 ng at 1 cell stage) at stage 17. (B) Western blot for pcdh8l and Rpn1 from CNC glycoproteins. Proteins were extracted from non-injected and MO13 injected CNC (Alfandari et al., 2003) and glycoproteins pulled down using Con-A-agarose beads. Western blot shows that MO13 at 10 ng reduces pcdh8l expression compared to non-injected. Error bars represent standard error to the mean (Mean ±S.E.M). One-way ANOVA was performed to determine statistical significance. Statistically significant at **p<0.01.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.26898.008
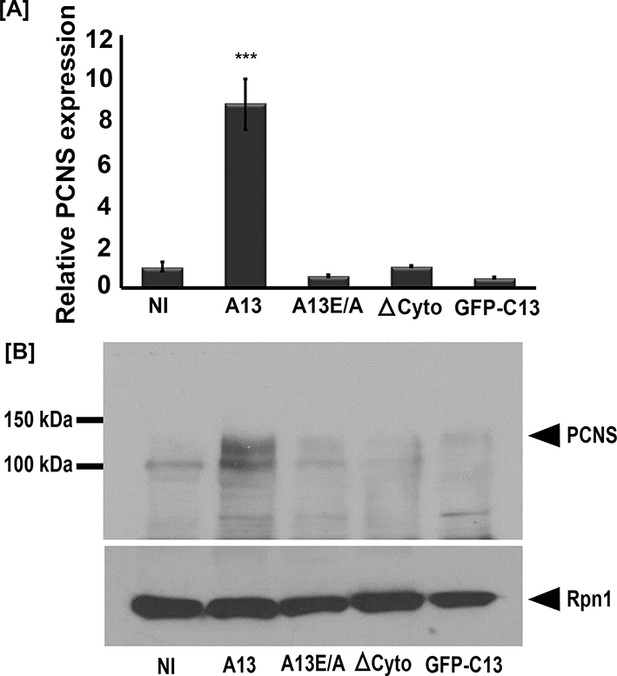
Full-length adam13 induces pcdh8l expression in naïve ectoderm (animal cap cells).
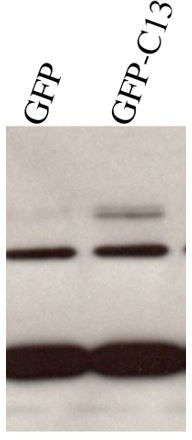
One-cell stage embryos were injected with mRNA encoding various forms of adam13 (1 ng). Animal cap (AC) explants were dissected at stage 9 and grown in 0.5X MBS until sibling embryos reached stage 18 to 20. mRNA and protein was extracted from AC for analysis of pcdh8l. (A) Quantitative real-time PCR from mRNA isolated from 10 animal caps. The relative level of pcdh8l expression was normalized to GAPDH. (B) Western blot for pcdh8l protein using mAb2F4. 30 AC were used to extract protein from embryos injected with various adam13 constructs. Western blot shows induction of pcdh8l by full-length adam13, whereas other constructs fails to induce pcdh8l expression. adam13 (A13), non proteolytic adam13 (A13E/A), adam13 lacking a cytoplasmic domain (ΔCyto), isolated adam13 cytoplasmic domain (GFP-C13). Error bars represent standard error to the mean (Mean ±S.E.M). One-way ANOVA was performed to determine statistical significance. Statistically significant at ***p<0.001.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.26898.011
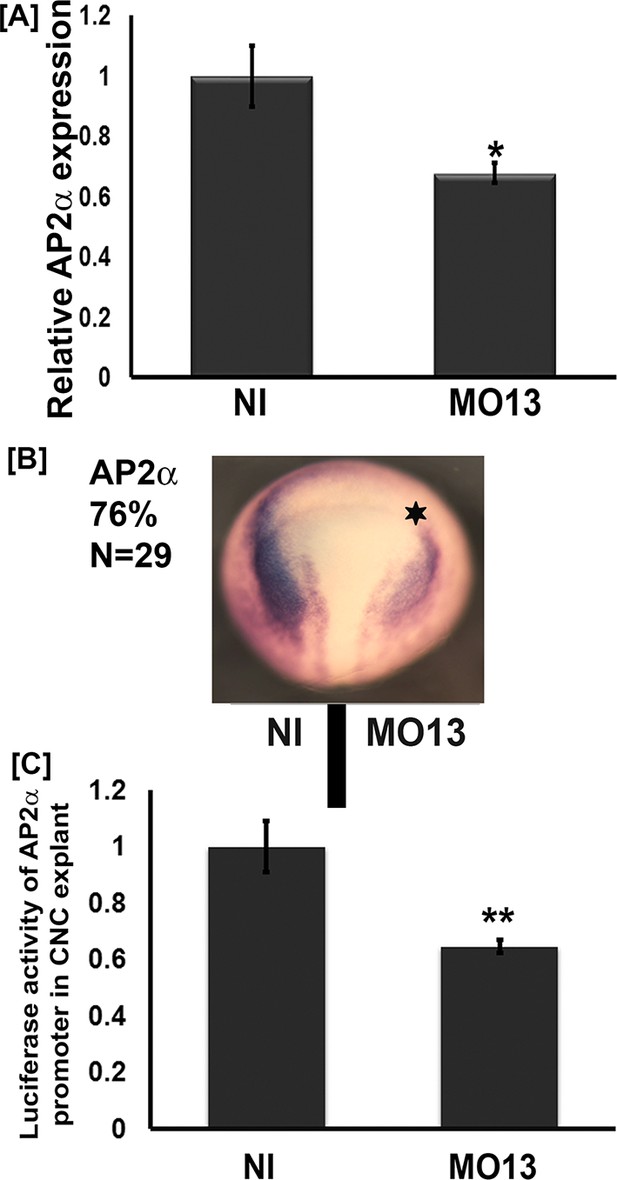
Adam13 knockdown reduces tfap2α expression.
(A) Relative mRNA expression of AP2α normalized to GAPDH (five embryos). Real-time PCR analysis shows a 40% decrease in AP2α expression in response to adam13 KD (B) Representative images of dorsal view of whole mount in situ hybridization with AP2α. MO13 (5 ng) was injected in one of the two blastomeres (star). The non-injected side serves as control. adam13 KD reduces AP2α expression in 76% of the embryos (N = 29). (C) Luciferase activity of AP2α promoter in cranial neural crest cells (CNC). One-cell stage embryos were injected with MO13 (10 ng). Control and KD embryos were further injected at the 8 cell stage with the AP2α:luciferase reporter (100 pg) together with pRL-CMV (10 pg) in one animal dorsal blastomere. CNC explants were dissected from stage 15–17 embryos and individual explants were lysed to measure luciferase activity. The normalized values from 10 individual explants (two biological replicates) were used for each measure. Error bars represent standard error to the mean (Mean ±S.E.M). One-way ANOVA was performed to determine statistical significance. Statistically significant at *p<0.05, **p<0.01.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.26898.013
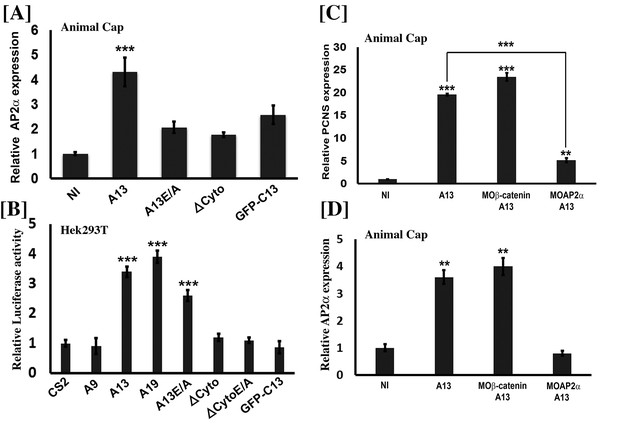
Adam13 induces pcdh8l via AP2α.
(A, C, D) Relative expression of AP2α and pcdh8l in naïve ectoderm (animal cap) by real-time PCR. One-cell stage embryos were injected with 1 ng of the various adam13 constructs and morpholinos and embryos were collected at stage 9 to dissect animal cap cells (AC). AC were grown in 0.5X MBS until sibling embryos reached stage 20 to 22. Messenger RNA was extracted from 10 AC for gene expression analysis. Expression was normalized using GAPDH and compared to the expression in non-injected AC (NI). (A) Real-time PCR data show that adam13 can induce AP2α by more than four fold. Both proteolytic activity and the presence of the cytoplasmic domain are essential for full activation. (B) Luciferase activity of AP2α promoter in Hek293T cells shows induction by adam13 and ADAM19 but not ADAM9. For each transfection the luciferase values were normalized to the Renilla values driven by the CMV promoter. In these assays, the absence of proteolytic activity (A13E/A) reduced AP2α induction only slightly, while the deletion of the cytoplasmic domain (ΔCyto) prevented the activity. (C) Induction of AP2α by adam13 was prevented by the KD of AP2α (10 ng of MOAP2α) but not β-catenin (20 ng of Moβ-catenin). (D) Expression of AP2α in response of adam13 also depends on AP2α but not β-catenin. Error bars represent standard error to the mean (Mean ±S.E.M). One-way ANOVA was performed to determine statistical significance. Statistically significant at **p<0.01, ***p<0.001.
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.26898.015
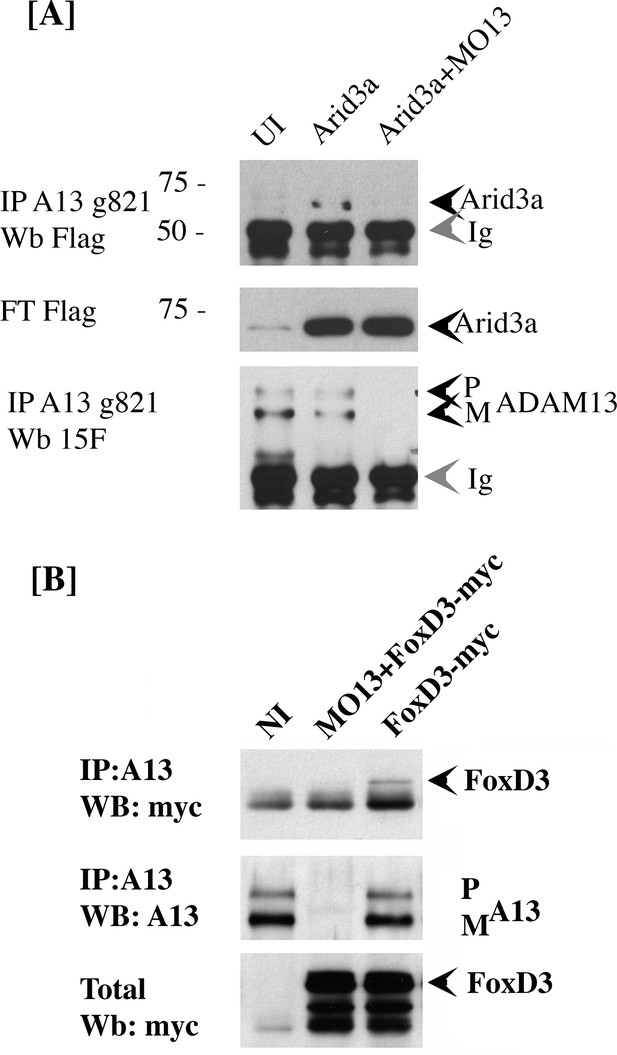
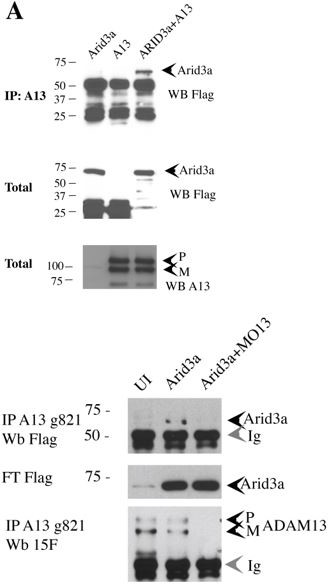
adam13 binds to arid3a and foxd3.
(A) Co-immunoprecipitation of arid3a-flag with adam13. Arid3a-flag mRNA were injected in one-cell stage embryos either alone or with the morpholino to adam13 (MO13.) Proteins were immunoprecipitated with a goat polyclonal antibody directed against the cytoplasmic domain of adam13 (g821, [Cousin et al., 2011]) and blotted with M2 to detect the Flag-tag of arid3a. The flow through was used to detect arid3a. The immunoprecipitation were re-probed using 6615F to detect adam13 (Alfandari et al., 1997). (B) Co-immunoprecipitation of adam13 and FoxD3 from embryos. FoxD3-myc mRNA was injected at the one-cell stage either alone or with MO13. Adam13 was immunoprecipitated using the goat polyclonal antibody to adam13 (g821), and the proteins were detected by western blot using either the myc antibody (9E10) or a rabbit antibody to adam13 (6615F). FoxD3 co-precipitated with endogenous adam13.
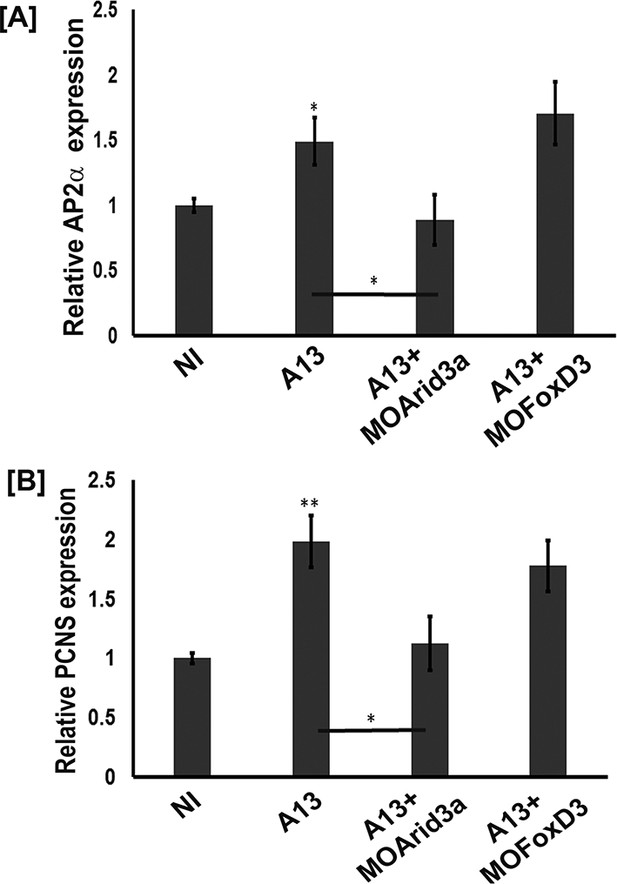
adam13 requires arid3a for induction of AP2α.
Relative expression of AP2α (A) and pcdh8l (B) in animal caps from embryos injected with adam13 alone or with a morpholino to arid3a (20 ng) or FoxD3 (20 ng). Animal caps were extracted at stage 17.
-
Figure 7—source data 1
Source data for Figure 7.
- https://doi.org/10.7554/eLife.26898.020
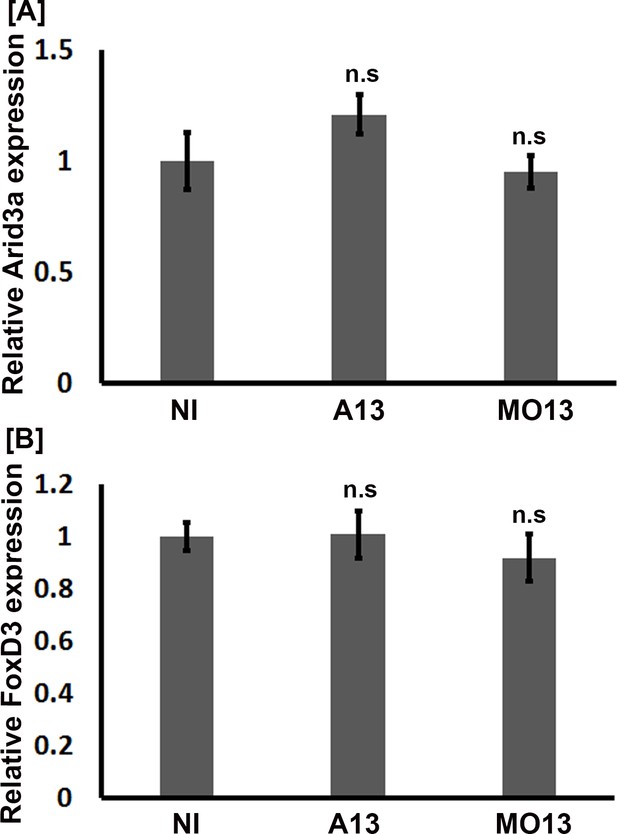
adam13 does not induce arid3a and FoxD3 expression in Naïve ectoderm.
Animal caps were dissected from stage 9 embryos injected with the various constructs or morpholinos as indicated. RNA was extracted from AC at stage 20. Graphs show relative mRNA expression normalized to GAPDH for (A) arid3a and (B) FoxD3. Error bars represent standard error to the mean for three or more independent experiments. One-way ANOVA was performed to determine statistical significance. Statistically significant at *p<0.05, ***p<0.005.
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1.
- https://doi.org/10.7554/eLife.26898.019
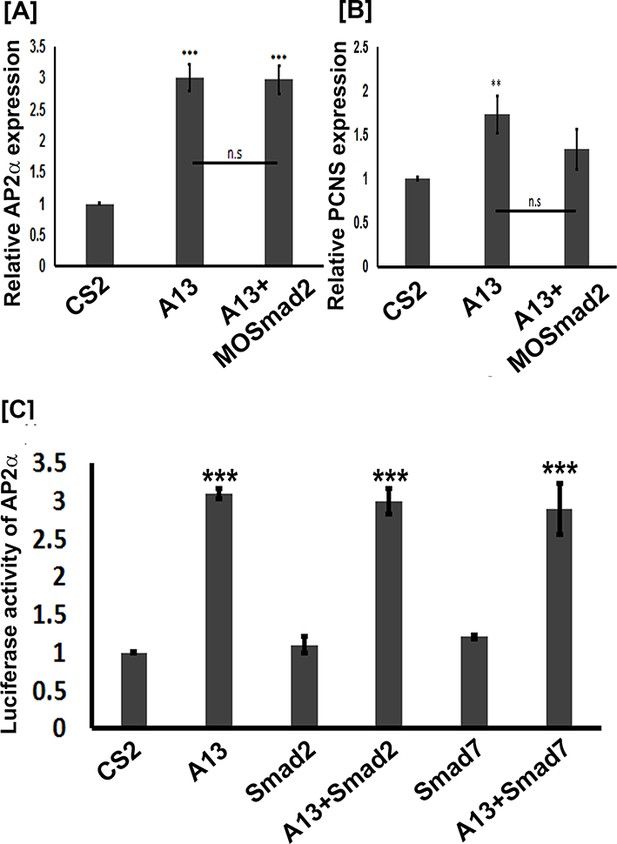
adam13 induction of Ap2α and pcdh8l does not involve Smad2.
(A–B) Relative expression of AP2α and pcdh8l in animal caps dissected from control embryos, or embryos injected with adam13 or adam13 and a morpholino to Smad2 (MOSmad2, 25 ng). (C) Luciferase assays in Hek293T cells show no induction of AP2α promoter activity by Smad2 (0.5 µg) or Smad 7 (0.5 µg). Smad2 does not increase adam13 activation of the AP2α promoter, while Smad7 does not reduce adam13 induction of AP2α promoter. Smad2 and Smad7 were transfected with or without adam13 (0.5 µg) along with AP2α promoter-luciferase and the pRL-CMV (100 ng, 10 pg). The ratio of luciferase to renilla was used to normalize each transfection to the empty vector control (CS2). Error bars represent standard error to the mean (Mean ± S.E.M). One-way ANOVA was performed to determine statistical significance. Statistically significant at **p<0.01, ***p<0.001.
-
Figure 8—source data 1
Source data for Figure 8.
- https://doi.org/10.7554/eLife.26898.022
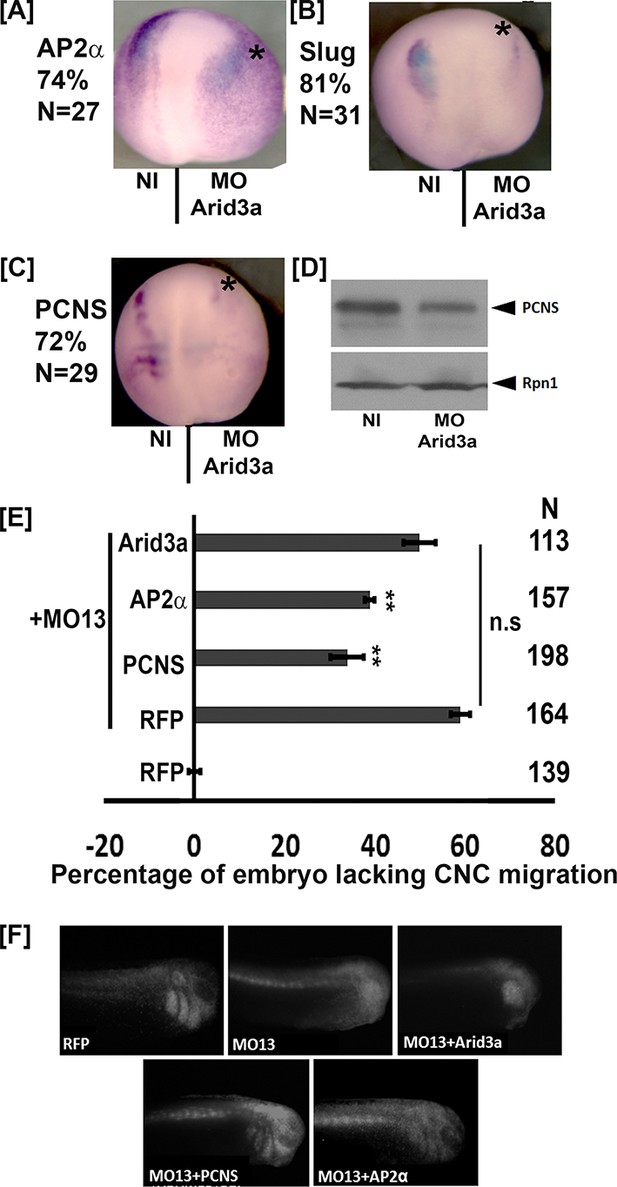
arid3a is critical for multiple gene expression in the CNC.
(A–C) Representative dorsal view of neurula treated by whole mount in situ hybridization with probes for tfap2α (AP2α), snai2 (Slug) and pcdh8l (PCNS). Eight-cell stage embryos were injected in one dorsal animal blastomere with the arid3a morpholino (5 ng, Asterisk). The percentage of embryos with reduced signal in the injected side is given in N is the total number of embryos obtained from each case. (D) Western blot using mAb2F4 to detect pcdh8l (PCNS). Glycoproteins from 25 embryos were extracted and purify on ConA-agarose beads. Rpn1 was used as a loading control. (E) Histogram representing the percentage of embryos lacking CNC migration. One-cell stage embryos were injected with MO13. Control or KD embryos were further injected at the 8 cell stage in one animal dorsal blastomere with mRNA for RFP, PCNS, AP2α or Arid3a. Observation of the RFP fluorescence at stage 24–26 reveals that the inhibition of migration by adam13 KD is partially rescued by pcdh8l and AP2α but not Arid3a. Error bars represent standard error to the mean (Mean ± S.E.M). One-way ANOVA was performed to determine statistical significance. Statistically significant at *p<0.05, **p<0.01. (F) Representative examples of injected embryos. Posterior to the left, dorsal is up.
-
Figure 9—source data 1
Source data for Figure 9.
- https://doi.org/10.7554/eLife.26898.024
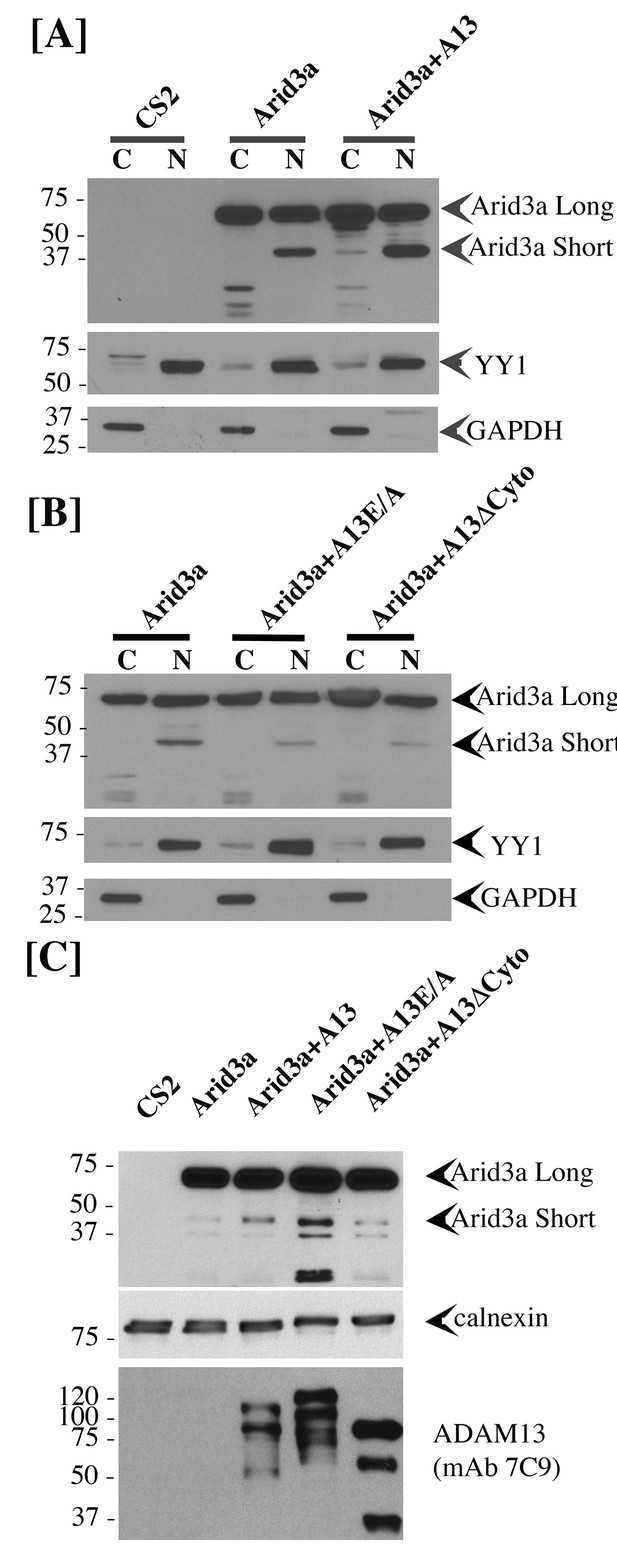
adam13 regulates arid3a post-translational modification.
Western blot from transfected Hek293T cells. (A–B) cytoplasmic (C) and nuclear (N) extracts from cells transfected with the empty vector (CS2), Arid3a-flag, adam13 (A13) or both. The blots were re-probed with the transcription factor YY1 as a nuclear marker and GAPDH as a cytoplasmic marker. The full-length Xenopus laevis Arid3a is detected at approximately 60 kDa (Arid3a Long). A shorter fragment is detected at about 40 kDa (Arid3a Short). (A) Co-transfection of adam13 with Arid3a increases the Arid3a protein level in the cytoplasm by 30% and the shorter fragment of Arid3a in the nucleus by five folds. (B) Co-transfection of Arid3a with the proteolytically inactive mutant adam13 (A13E/A) or the mutant lacking the cytoplasmic domain (A13ΔCyto) does not increase the shorter fragment in the nucleus. (C) Membrane extract from Hek293T cells transfected with Arid3a-flag and the adam13 constructs. Co-transfection of Arid3a with adam13 increases the intensity of the 40 kDa Arid3a fragment. This is not observed in the absence of the adam13 cytoplasmic domain. In contrast, a much more significant increase is observed when Arid3a is co-transfected with the A13E/A mutant.
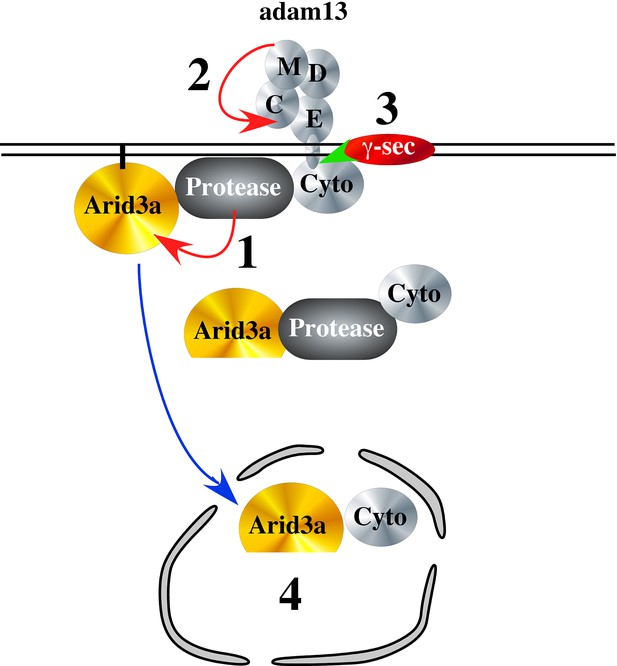
Hypothetical model of adam13 function: Adam13 at the membrane may associate with a cytoplasmic protease that cleaves Arid3a (1) to generate the short form seen in Figure 10.
Arid3a is localized to lipid raft via its palmytoylation. Once adam13 cleaves itself in the cysteine rich domain (2), gamma secretase cleaves the cytoplasmic domain of adam13 (3), releasing the complex that can translocate in the nucleus to activate tfap2α transcription.
Tables
| Target | Sequence 5’−3’ |
|---|---|
| xltfap2α Forward | ATAACAATGCGGTGTCGTCCCTCT |
| xltfap2α Reverse | AGAGCCTTCTCTGGACTTCTGCAA |
| xlarid3a Forward | GGAGGCTTGGTGGAAGTTATTA |
| xlarid3a Reverse | ACTGAGTGCGCAGAGTAAAG |
| xlGAPDH Forward | TTAAGACTGCATCAGAGGGCCCAA |
| xlGAPDH Reverse | GGGCAATTCCAGCATCAGCATCAA |
| xlpcdh8l Forward | TCTCAACTCGTGCTCAAATC |
| xlpcdh8l Reverse | CCTCTGCTGACCCATTATTC |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26898.028