Mouse embryonic stem cells can differentiate via multiple paths to the same state
Figures
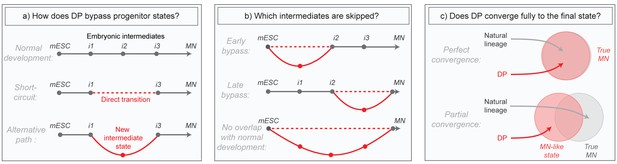
Summary of core research questions.
(a) Direct programming could in principle skip progenitor states in one of two ways. It may either transition from early states directly into later states through a ‘short-circuit’ of the natural lineage, or it might utilize an alternative path involving new and potentially abnormal or unstable intermediate states. In the schematic, a conceptual depiction of the natural lineage is shown first, followed by two lineages that bypass the i2 intermediate from the normal lineage, either through short-ciruit or through a new alternative intermediate. (b) DP might diverge from the natural lineage only briefly, or it might access an entirely distinct path. In the schematic, three different possibilities are shown: (i) DP bypasses specific early intermediates, and then converges to the final state through a conserved seqeunce of terminal cell state transitions; (ii) DP transitions through conserved early stages of differentiation, before diverging into an alternate path that converges separately to the final state; (iii) DP takes an entirely distinct path with no resemblance to the natural lineage. Dashed lines represent a potential direct transition while solid lines represent an alternate path (as shown in the left panel). (c) It is possible that the price paid for taking an alternate differentiation path during direct programming is that cells retain subtle gene expression defects, thus converging only partially to the desired final state. Alternatively, convergence could be near perfect which would imply that differentiation into mature cell states is at least partially history independent. In the schematic, circles are an abstract representation of gene expression state and are used to represent the extent of overlap between MNs generated by either DP or by the natural lineage.
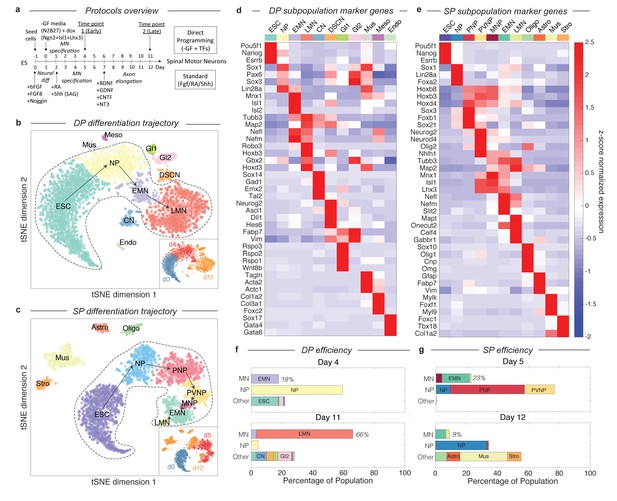
Dissection of DP and SP motor neuron differentiation strategies using InDrops single cell RNA sequencing.
(a) Summary of the direct programming (DP) and standard protocol (SP) differentiation strategies. (b–c) tSNE visualization of single cell RNA sequencing data from each differentiation strategy. Timepoints are shown inset. Cell state clusters are color coded and annotated with their identities: ESC = embryonic stem cell; NP = neural progenitor; PNP = posterior neural progenitor; PVNP = posterior and ventral neural progenitor; MNP = motor neuron progenitor; EMN = early motor neuron; LMN = late motor neuron; CN = cortical neuron; DSCN = dorsal spinal cord neuron; Gl1 = glia type 1; Gl2 = glia type 2; Mus = muscle; Meso = mesoderm; Endo = endoderm; Oligo = oligodendrocyte; Astro = astrocyte; Stro = stromal. Arrows inside the bounded area indicate the hypothesized cell state progression during MN differentiation for each method. (d–e) Expression heatmap of marker genes used to identify each subpopulation. Colors and annotations for the subpopulations are the same. (f–g) Subpopulation abundances for each protocol over time. DP has significantly higher MN production efficiency than SP (66% vs. 9%) at the late timepoint. MN = EMN or LMN; NP = NP, PNP, or PVNP; Other = everything else. Colors and labels match b-e).
-
Figure 2—source data 1
Summary of criteria used to annotate DP cell states.
Table indicates the expected expression level of key marker genes for each cell state. Hi = high expression, Lo = low expression, ‘-’=no expression. If an expression level is not specified for a given state-gene pair, then that gene was not a component of the criteria used to annotate state. Real expression values are shown in Figure 2D, and Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.26945.007
-
Figure 2—source data 2
Summary of criteria used to annotate SP cell states.
Table indicates the expected expression level of key marker genes for each cell state. Hi = high expression, Lo = low expression, ‘-’=no expression. If an expression level is not specified for a given state-gene pair, then that gene was not a component of the criteria used to annotate state. Real expression values are shown in Figure 2E, and Figure 2—figure supplement 2.
- https://doi.org/10.7554/eLife.26945.008
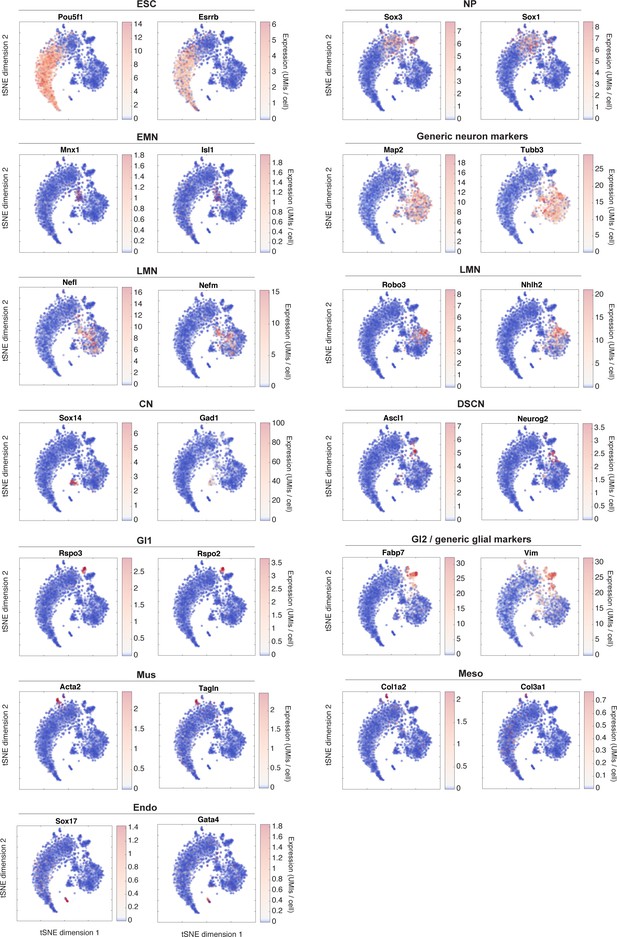
tSNE visualization of marker genes for DP subpopulations.
This figure relates to main text Figure 2. It shows the raw expression of sample marker genes that were used to identify each subpopulation. Two marker genes are shown per state.
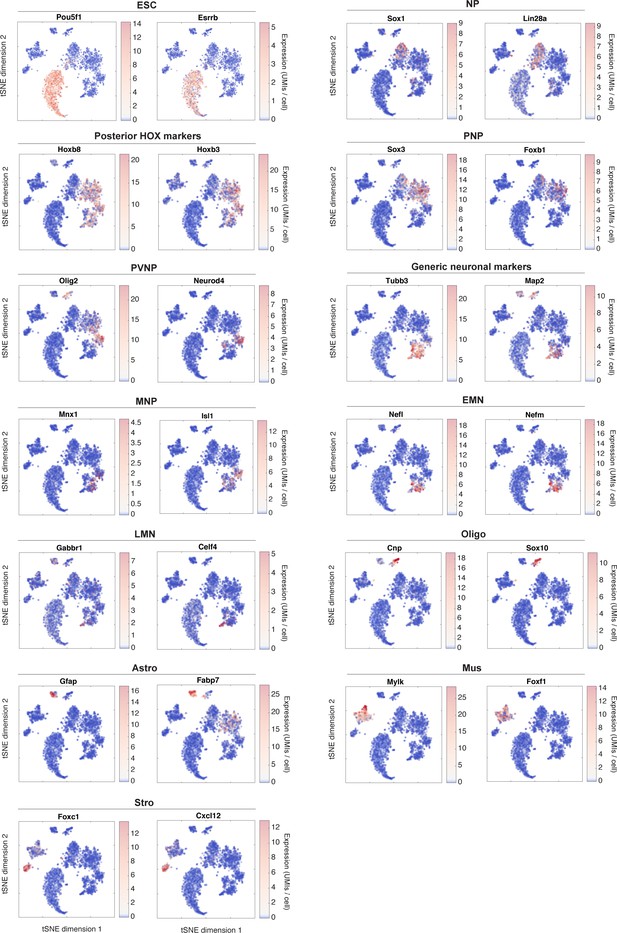
tSNE visualization of marker genes for SP subpopulations.
This figure relates to main text Figure 2. It shows the raw expression of sample marker genes that were used to identify each subpopulation. Two marker genes are shown per state.
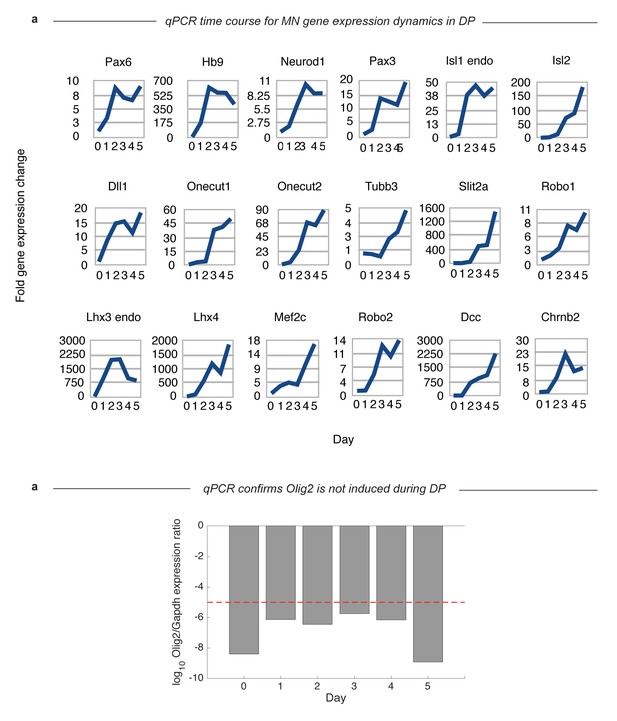
qPCR validation that MN gene expression in DP follows the expected dynamics and does not involve Olig2 induction.
(a) Measurement of the bulk population gene expression over time helps to confirm that the trajectory inferred from single-cell analysis matches the true dynamical events of our system. These results confirm that key MN differentiation markers are induced on a real timescale that matches their ordering in our inferred trajectory. Terminal genes such as Mnx1 and Tubb3 are upregulated immediately after induction of early progenitor genes such as Sox1. The values plotted are shown in units of fold change relative to their expression in day 0 mESC cultures. (b) Plot shows that Olig2 is expressed >= 10∧6 times lower than Gapdh during DP. Red line indicates the expected ratio if 0.1% of the population expressed one molecule per cell; the observed ratios are below even this low bound.

DP and SP induce distinct differentiation paths to the motor neuron state.
(a) Visualization of differentiation paths for both protocols using SPRING reveals two paths to the same state. DP and SP overlap during early neural commitment, but then bifurcate and converge to the same final MN state separately. Reds = direct programming path; Blues = standard protocol path; arrows indicate hypothezed differentiation trajectories. Cell states are colored and labeled by according to their definition in Figure 1 for comparison. (b) Gene expression of key marker genes along each differentiation path confirms exit from the pluripotent state (Oct4) and progression towards the MN state (Tubb3, Stmn2) for both protocols. Only the SP upregulates Olig2 and Nkx6-1, which mark important MN lineage intermediates in the embryo; this occurs following the bifurcation of both paths. Expression in cells from each sample are colored using either a blue (ESC), red (DP), or green (SP) colormap to allow tracking of each path separately. (c) Pairwise cosine similarity of cell state centroids. Note early and late similarity of states, but prominent differences during intermediate state transitions. (d) Every individual cell of the DP trajectory was assigned to its most similar SP state using a maximum likelihood method. DP cells map to early and late but not intermediate states of the SP cell state progression.
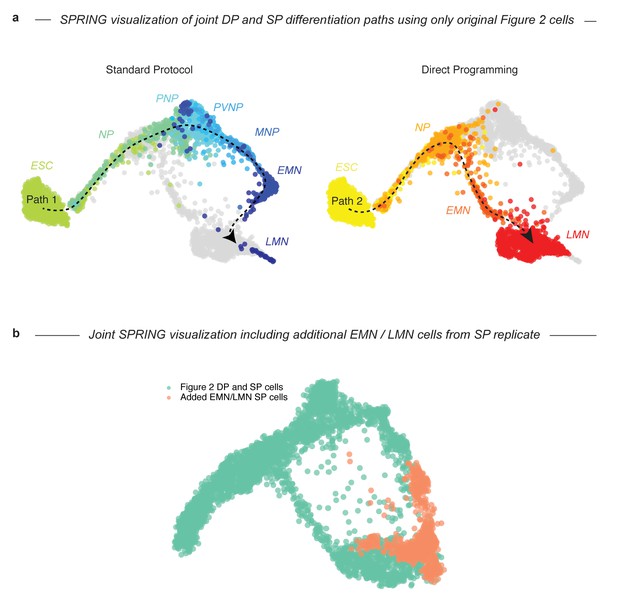
Consolidation of DP vs SP end-state comparisons with additional SP EMN and LMN cells does not distort joint topology.
In our initial InDrops map of the SP, we found a low yield of the most mature EMN and LMN states in the SP (Figure 2C). To ensure that low representation of the SP end states did not interfere with our ability to make statistically rigorous comparisons between methods, we integrated additional EMN and LMN cells from a replicate SP experiment into our analyses. This figure serves as a control that shows that the topology of our joint SPRING visualization comparing DP and SP does not change from the case where we compare only cells from the original experiment (a), to when we compare the DP and SP using added EMN and LMN cells from the replicate SP experiment (b). As expected, the added cells specifically improve representation of terminal SP populations.
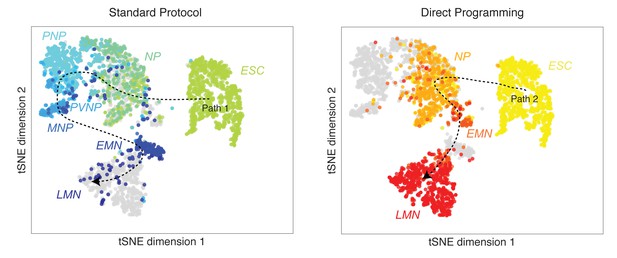
Joint visualization of DP and SP differentiation paths by tSNE.
Like SPRING, tSNE shows similarity between early and late states, but it is ambiguous with respect to the continuous global topology of either path. Between DP and SP, tSNE confirms that ESC, NP, and LMN states between DP and SP are similar. Moreover, SP intermediate states (PNP, PVNP, and MNP) do not overlap with the DP intermediate state (EMN). However, since tSNE emphasizes local similarity relationships the overall joint topology, as revealed by SPRING in Figure 3A, is in this case ambiguous.
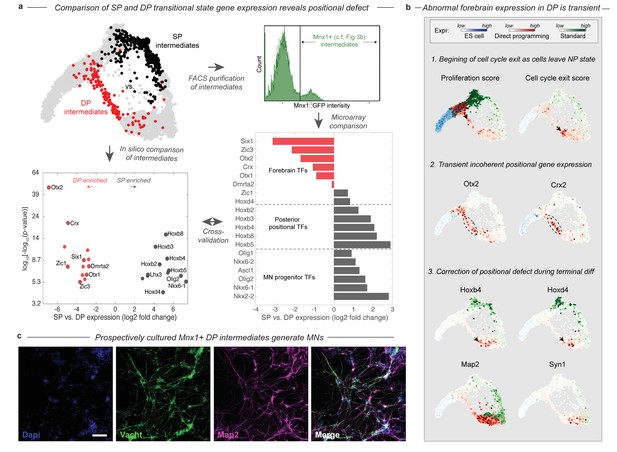
During DP cells transition through an abnormal intermediate state with a forebrain gene expression defect.
(a) The DP (red, EMN from Figure 2) and SP (black, PVNP + MNP + EMN from Figure 2) differentiation paths diverge into distinct intermediate states following early neural commitment. These distinct populations were compared by in silico differential expression analysis using single cell data (bottom left), or by FACS purification of the intermediate populations using and Mnx1::GFP reporter cell line followed by microarray analysis (bottom right). Figure 3b. shows that Mnx1 expression localizes to the distinct intermediate populations of both protocols, making it an appropriate target for their purification. Differential expression analysis revealed a total of 26 differentially expressed TFs between the intermediate populations (>6 fold differentially expressed, p<0.001), of which (61%) were involved in either forebrain positional identity (Otx2, Crx, Zic1, Six1, Dmrta2, Otx1, and Zic3) and enriched in DP, or in posterior spinal identity (Hoxb8, Hoxb3, Hoxb2, Hoxb4, Hoxb5, Hoxd4, Lhx3, Olig2, and Nkx6-1) and enriched in SP. The independent microarray comparison confirmed these differences with only one exception - Zic1. It also revealed that the SP intermediates are enriched for additional spinal neural progenitor genes including Ascl1, Nkx6-2, Olig1, and Nkx2-2. (b) Before forebrain genes are expressed, DP cells have a high proliferation score and low cell-cycle exit score (computed as the sum of a panel of cell-cycle-associated or tumor supressor genes respectively). They reduce cell cycle gene expression as they enter the abnormal transitional state, and upregulate forebrain genes including Otx2 and Crx. This abnormal forebrain expression is shut off as cells exit the transitional state into the final MN state, characterized by Map2 and Vacht expression. The final transition into a MN state is also accompanied by upregulation of posterior positional genes including Hoxb4 and Hoxd3, thus correcting the transient forebrain positional expression defect. Expression in cells from each sample are colored using either a blue (ESC), red (DP), or green (SP) colormap to allow tracking of each path separately. (c) Mnx1+ cells were isolated at day 3 of differentiation and cultured to determine their fate potential. By 12 days after replating most cells expressed motor neuron markers Map2 and Vacht. Scale bar = 100 um.
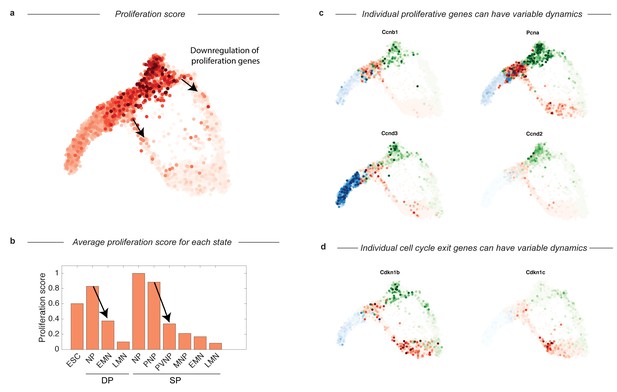
Cell-cycle-associated gene expression decreases as cells enter the DP and SP intermediate states and then progressively mature.
This figure relates to main text Figure 4B. (a) Cell cycle gene expression score overlayed on SPRING trajectory shown in main text. The score is the summed expression of a panel of 21 cell-cycle related genes. Cell cycle expression is high in ES cells and neural progenitors from both protocols, but drops sharply upon entry into the transitional states preceding terminal differentiation. (b) Shows the average proliferation score along both DP and SP trajectories computed by state (and max normalized). Arrows indicate the same transition as the arrows shown above in panel a. (c–d) Example raw expression data for representative individual cell cycle or tumor suppressor/cell cycle exit genes. Some genes appear to be enriched in specific states, such as Ccnd3 in ES cells and Ccnd2 in neural progenitors from the SP. Expression in cells from each sample are colored using either a blue (ESC), red (DP), or green (SP) colormap.

Both DP and SP differentiation trajectories approach the transcriptional state of primary MNs (pMNs), but DP does so with higher precision.
(a) tSNE visualization of 874 single cell transciptomes from FACS purified Mnx1+ MNs from embryos reveals heterogeneity within this population. To make comparisons between DP and SP with pMNs we used only the subset of Mnx1:GFP+ primary cells in a bona-fide MN state. See Figure 5—figure supplement 1 for marker gene expression in each population. (b) Comparison of average gene expression profiles for cell states along the DP and SP trajectories with pMNs. In both methods similarity increases as differentiation proceeds. Late DP states are the most similar to embryonic MNs. (c) Projection of the reference E13.5 pMNs into the visualization from Figure 3 revealed that pMNs closely associate with the terminal states of both DP and SP (i). Close examination of the terminal populations (EMN, LMN) from DP and SP compared to pMNs reveals heterogeneity representing state subtypes (ii). At a single cell level DP LMNs were the most closely associated with E13.5 pMNs; 64% of DP LMNs had at least 1 pMN nearest neighbor out of its most similar 50 cells compared to 6% for SP LMNs (iii). The subtypes present within terminal DP and SP populations could be annotated using marker genes. DP and SP EMNs express progenitor genes including Mnx1, along with Nkx2-2 and Nkx6-1 in SP only. The major SP LMN outgroup expressed Gata3, indicating a hindbrain identity. Both DP LMNs and pMNs shared expression of the terminal MN differentiation gene Ebf2. (d) Systematic pairwise differential gene expression analysis between terminal DP and SP states and pMNs. Each panel is a volcano plot of differentially expressed transcription factors. Both DP and SP LMNs show limited gene expression differences to pMNs. The dominant differences are positional, with DP and SP LMNs lacking the most posterior Hox genes. Other expression differences are explained by differences in terminal state subtypes as shown in c). Differentially expressed genes were filtered for TFs with a corrected p-value<0.001, expression ratio > 4, and minimum expression of 1umi/cell average.
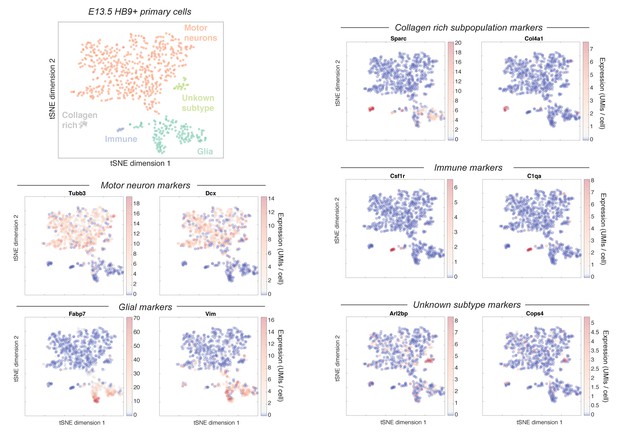
Heterogeneity within the Mnx1+ E13.5 primary motor neuron population.
This figure shows the raw expression of a set of marker genes that were used to identify the subpopulations indicated in main text Figure 5A. Two markers are shown per subpopulation, with their expression intensity overlaid on the original tSNE embedding.

Validation that DP cells become functional MNs despite their abnormal differentiation trajectory.
(a) Immunostaining of MN markers in DP MNs confirming expression and correct subcellular localization of Tubb3, Map2, VACht, Isl1, and Hb9. DP MNs also: b) can fire single or multiple action potentials upon stimulation, (c) show sodium and potassium channel currents, and d) are responsive to multiple MN neurotransmitters - AMPA, kainate, GABA, and glycine. (e) Co-culture experiments show that DP MNs can induce clustering of acetylcholine receptors on muscle myotubes (indicated by α-BGT staining) and induce their contractions. The induced contractions are dependent on MN activity as they can be blocked by addition of the acetylcholine antagonist curare.
Contractions induced by DP MNs in C2C12 myotubes.
https://doi.org/10.7554/eLife.26945.017Contractions induced by DP MNs in ES-MyoD myotubes.
https://doi.org/10.7554/eLife.26945.018Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26945.019





