Dissection of specific binding of HIV-1 Gag to the 'packaging signal' in viral RNA
Figures
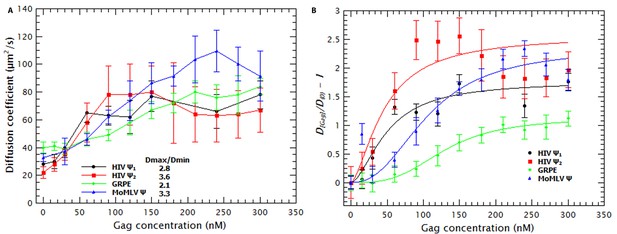
Binding of Gag collapses the RNA.
(A) The diffusion coefficient (D) of the Cy5-labled RNAs increases with increasing Gag concentration. The ratio of the maximum diffusion coefficient of bound (Dmax) and free RNA (Dmin) quantifies the degree of RNA collapse. Ψ1, monomeric Ψ; Ψ2, dimeric Ψ. (B) The normalized ratio of the maximum diffusion coefficient of bound (D[Gag]) and free RNA (Dmin) as a function of Gag concentration is best fit with a simple cooperative model (lines). The quality of the RNAs was tested by running denaturing gels (see Figure 1—figure supplement 1a) while native gels were used to verify that the dimerization protocol did not produce large aggregates (see Figure 1—figure supplement 1b). Sedimentation velocity and FCS were used to verify that there were no detectable Gag (Figure 1—figure supplement 2) or RNA (Figure 1—figure supplement 3) aggregates due to freezing/thawing cycles. A comparison of the distribution of the diffusion coefficients of the HIV Ψ that had been thermally annealed to produce either monomers or dimers is shown in Figure 1—figure supplement 4. Figure 1—figure supplement 5 shows that addition of Proteinase K to Gag/RNA complexes decreases the D to that of free RNA.
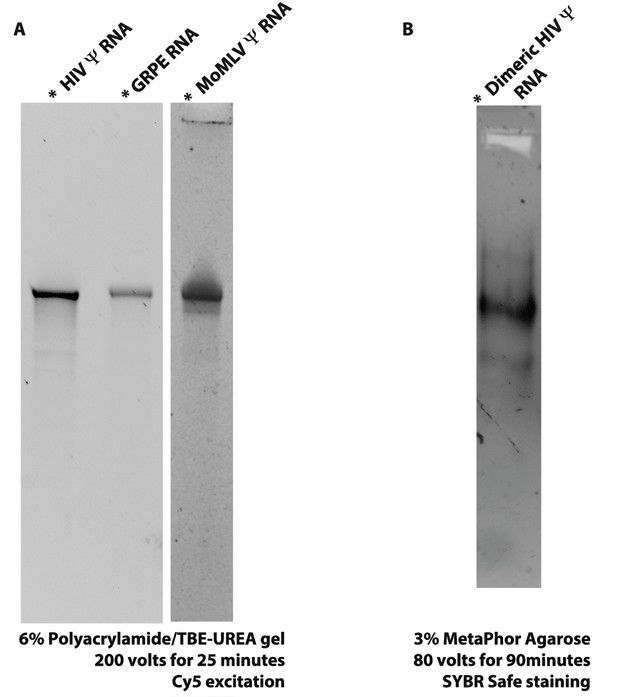
The quality of the RNAs was determined by gel electrophoresis.
(A) 6% polyacrylamide/TBE-UREA (i.e., denaturing) gels were used to verify the quality of the RNAs after Cy5 labeling. (B) The HIV-1 Ψ-Cy5 was electrophoresed in a 3% MetaPhor Agarose (i.e., non-denaturing) gel in TAE buffer to determine if the RNA dimerization protocol results in aggregation. These results are consistent with the FCS data (see Figure 1—figure supplement 4).
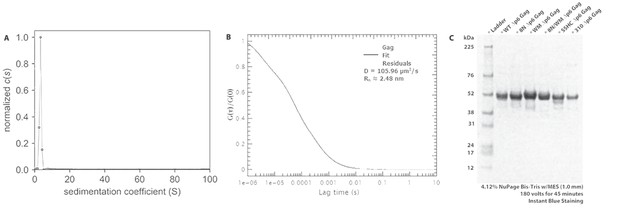
Freezing/thawing cycles of Gag does not yield protein aggregation.
(A) A solution of 2 μM Δp6 Gag was diluted from a stock solution of 20 μM, after freezing at −80°C, and was analyzed by sedimentation velocity. The c(s) profile is consistent with a reversible monomer-dimer self-association and shows a peak centered at 3.25 s, with 97% of all signal between 0.5–8.0 s. (B) The normalized autocorrelation function of AlexaFluor 487-labeled Δp6 Gag shows that there are no detectable aggregates; the diffusion coefficient agrees with a single-species with a Rh of 2.5 nm. The red line is the residual plot of the fit of the autocorrelation function. (C) 4–12% NuPAGE SDS-PAGE gel comparing WT Δp6 Gag (lane 2) and all the mutant proteins: 8N, WM, 8N/WM, SSHC and 310 Gag (lanes 3 through 7 respectively).
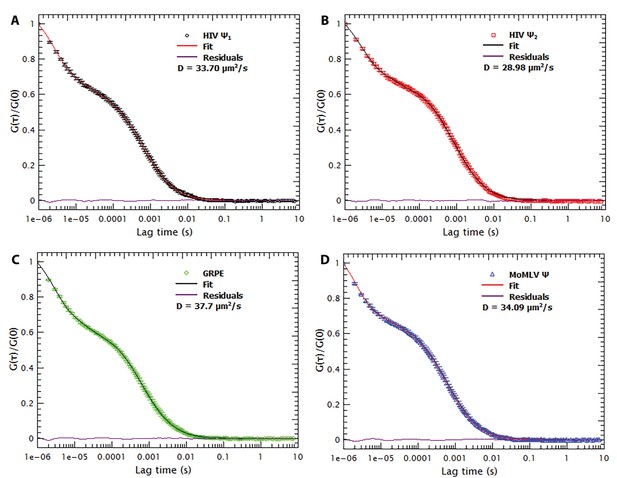
Freezing/thawing cycles and thermal annealing of the RNAs do not result in RNA aggregation.
The normalized autocorrelation functions for Cy5-labeled monomeric Ψ, dimeric Ψ, GRPE and MoMLV Ψ (A, B, C and D, respectively) shows that freezing/thawing cycles as well as the thermal annealing procedure does not produce large scale aggregates. Such aggregates would appear at lag times equal or larger than 0.01 s. The red line is the residual plot of the fit of the autocorrelation function.
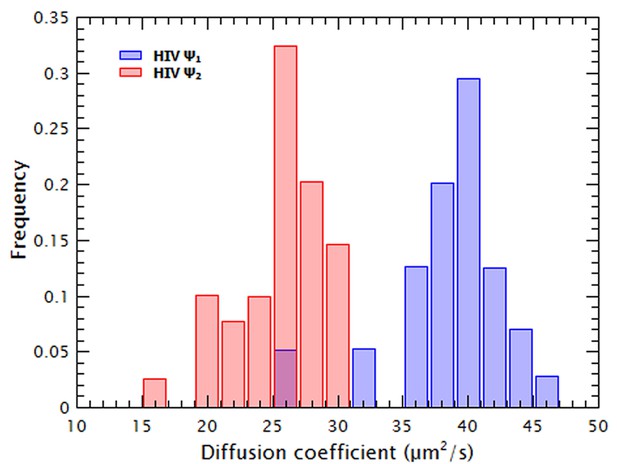
The thermally annealed HIV-1 Ψ dimers can be resolved from monomers by FCS and are stable at 15 nM.
The diffusion coefficient of HIV-1 Ψ RNA was measured after using a protocol to promote the formation of monomers (Ψ1, red bars) or dimers (Ψ2, blue bars). These frequency distributions of the diffusion coefficients of the two RNA species show that at the concentrations used in these experiments the dimeric RNA is stable and that at least 95% of the ‘dimeric’ RNA has a diffusion coefficient consistent with a dimeric RNA.
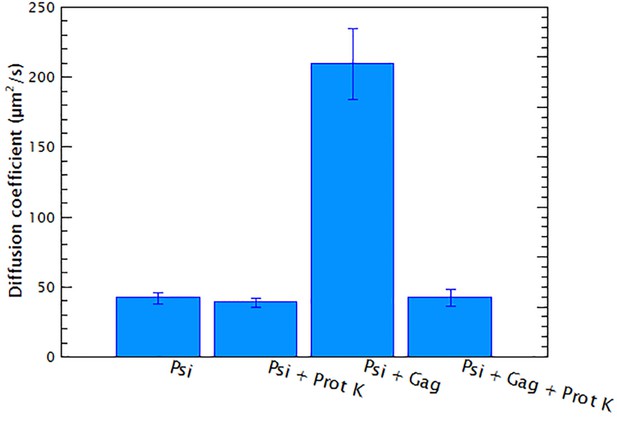
Proteolytic digestion of Gag/RNA complexes recovers the diffusion coefficient of the RNA.
15 nM of Cy5-labeled monomeric HIV-1 Ψ was mixed with 400 nM of HIV-1 Gag following the standard protocol described in Materials and methods. The diffusion coefficient of the RNA/Gag complex (Psi + Gag) and of pure RNA (Psi) were measured. After the measurements 1 μg of Proteinase K was added to each sample and the reaction mixtures were incubated for 1 hr at 37°C. The diffusion coefficients of the digested samples were measured again (Psi + Prot K and Psi + Gag + Prot K, respectively). This experiment shows that the increase in the diffusion coefficient, upon addition of Gag, is not due to RNA degradation and that Gag-induced RNA collapse is reversible.
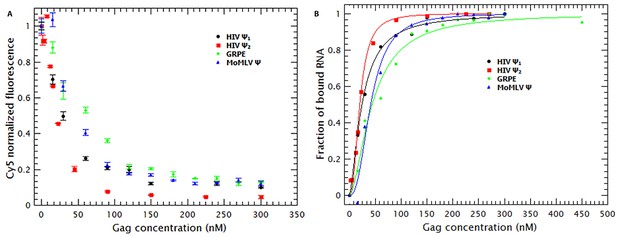
Binding of Gag quenches Cy5; quenching is a metric of binding.
(A) The fluorescence intensity of Cy5 decreases with increasing Gag concentration. (B) Binding plots of wt Gag with different RNAs, derived from quenching data in A. The binding plots are best fit to a cooperative model (lines). The binding plots show that the dimeric HIV-1 Ψ (HIV Ψ2) has the highest binding affinity. However, the difference in the binding affinity between this RNA and the other RNAs is minimal. Figure 2—figure supplement 1 shows a representative comparison of three different binding models; cooperative, multiple non-cooperative and non-cooperative binding.
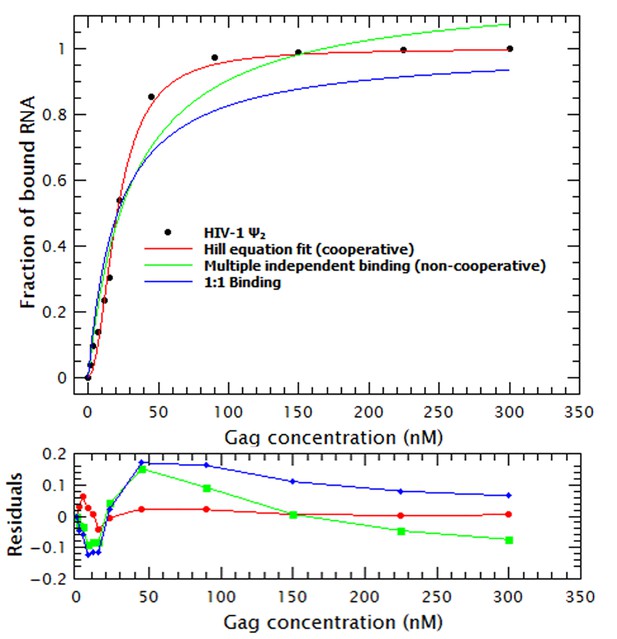
Binding of Gag to RNA is cooperative.
This binding plot of WT Gag with the dimeric HIV-1 Ψ is a representative example showing that a cooperative binding model (red line), using the Hill equation (Equation 7), describes the experimental data better than a non-cooperative model (green line) or a 1:1 binding mode (blue line).
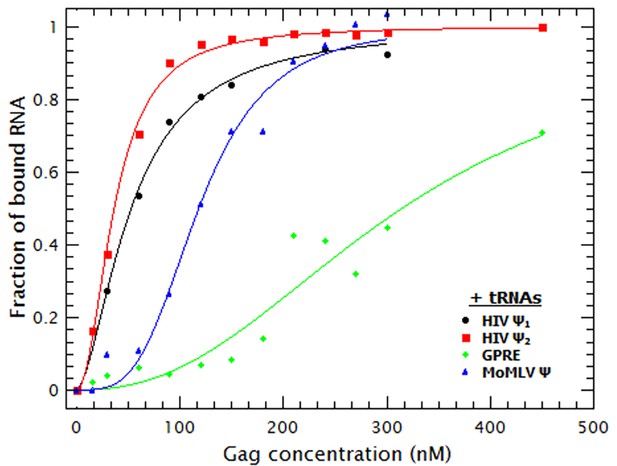
Addition of a competitor RNA reveals binding specificity.
Addition of a large excess of yeast tRNAs (≈120 moles of tRNA per mole of RNA) greatly increases the difference in the apparent affinity between that for the dimeric HIV-1 Ψ (Ψ2) (red sequares) and that for the other RNAs. The data was fitted with a cooperative binding model (solid lines). Figure 3—figure supplement 1 shows how both the diffusion coefficient of the RNAs and the Cy5 quenching data are affected by the presence of tRNAs.
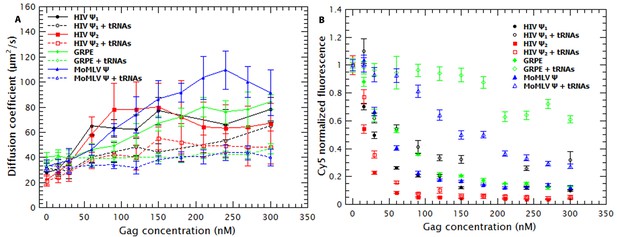
The presence of tRNAs increases the size of the Gag/RNA complexes.
(A) The increase in the diffusion coefficient of the RNAs in the Gag titrations is diminished by addition of tRNA. (B) The quenching data shows that the addition of tRNAs reduces the binding of Gag to the GRPE and MoMLV Ψ RNAs (green and blue symbols). The degree of quenching for the dimeric HIV-1 Ψ (red symbols) is almost independent of the presence of tRNAs.
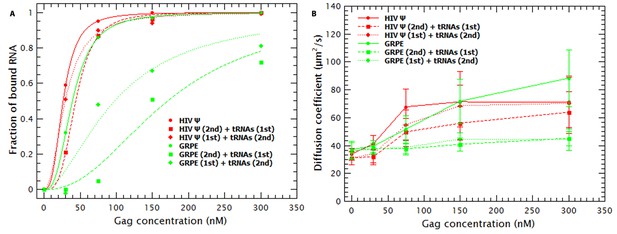
Hysteresis in Gag/RNA binding.
(A) The presence of excess tRNAs and the order in which the tRNAs are added to the reaction mixture with the monomeric HIV-1 Ψ have a minor impact in the binding plots (red data). In contrast, with GRPE RNA the order of addition, as well as the presence of excess tRNAs, has a large influence on the binding isotherm. (B) Addition of tRNAs after mixing HIV-1 Ψ and Gag (red diamonds) only blocks RNA collapse at low Gag concentrations. tRNAs block GRPE collapse independent of the order in which they are added (green squares and diamonds).
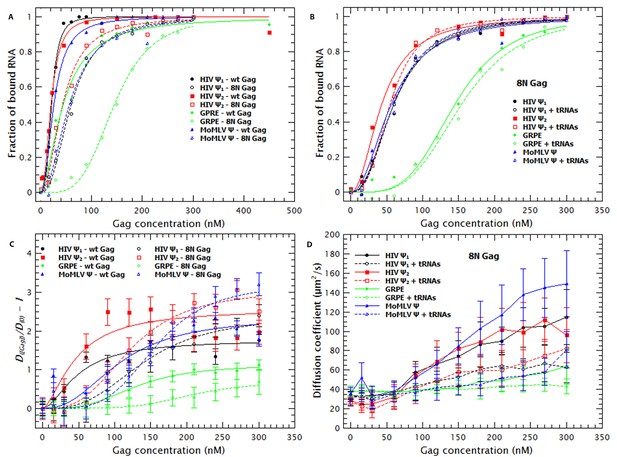
The Matrix domain of Gag contributes to non-specific binding but has no major impact on RNA collapse.
(A) Side-by-side comparison of the binding plots for WT (solid symbols) and 8N Gag (open symbols) to monomeric Ψ (circles), dimeric Ψ (squares), GRPE (diamonds) and MoMLV Ψ (triangles). Reducing the charge in MA affects the binding affinity for these RNAs. (B) The comparison of the binding plots for 8N Gag in the absence (solid symbols) and presence (open symbols) of a ≈120-fold molar excess of tRNAs. (C) Comparison of the normalized diffusion plots for the different RNAs upon binding of WT (solid symbols) and 8N Gag (open symbols). The lines are a cooperative fit of the Gag-induced RNA collapse. (D) As with WT Gag (solid symbols), the presence of excess tRNAs increases the size of the 8N Gag/RNA complexes (open symbols).
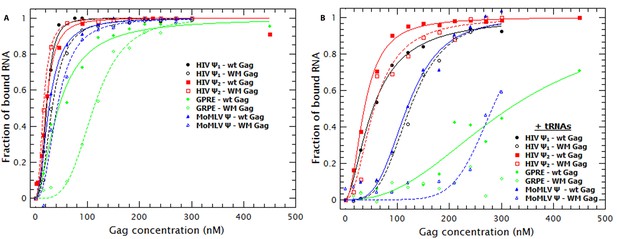
Non-specific binding is greatly decreased by decreasing the strength of Gag-Gag interactions.
(A) Comparison of the binding plots of WT (closed symbols) and WM Gag (open symbols). While the binding affinity for the monomeric and dimeric HIV-1 Ψ (black and red symbols, respectively) as well as the MoMLV Ψ (blue triangles) is not greatly affected by the WM mutation, binding to the GRPE RNA (green diamonds) is significantly impaired. (B) In the presence of tRNAs binding to the GRPE (green diamonds) and MoMLV Ψ (blue triangles) is greatly impaired while binding to the dimeric HIV-1 Ψ (red squares) suffers only a minimal decrease in binding affinity. Figure 6—figure supplement 1 compares the degree of RNA collapse by WT and WM Gag; RNA collapse by the mutant protein is impaired.
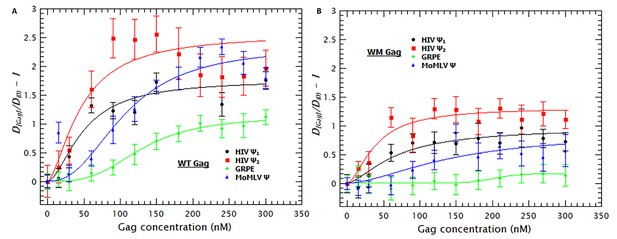
Reducing Gag-Gag interactions decreases the degree of RNA collapse.
Side-by-side comparison of the degree of RNA collapse by WT (A) and WM Gag (B). Panel A is the same as Figure 1B. With the WM mutation, which reduces Gag-Gag interaction, the degree of RNA collapse is severely impaired. The solid lines are fits of the experimental data using a cooperative model.
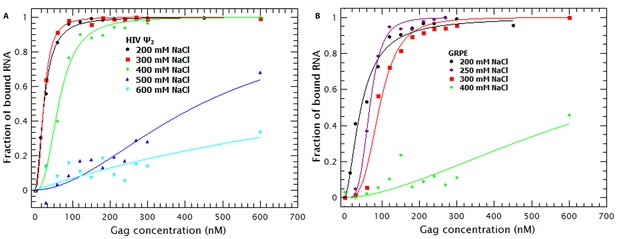
Binding of Gag to dimeric HIV-1 Ψ has a larger non-electrostatic component than binding to GRPE RNA.
(A) Binding of WT Gag and the dimeric HIV-1 Ψ (Ψ2) was measured at various ionic strengths. Increasing the salt concentration from 0.2 to 0.4 M NaCl had a minor effect on the binding affinity of the dimeric Ψ. Binding to dimeric HIV-1 Ψ (Ψ2) was detectable as high as 0.6 M NaCl. (B) The highest salt concentration at which we could measure binding to the GRPE RNA was 0.4 M NaCl.
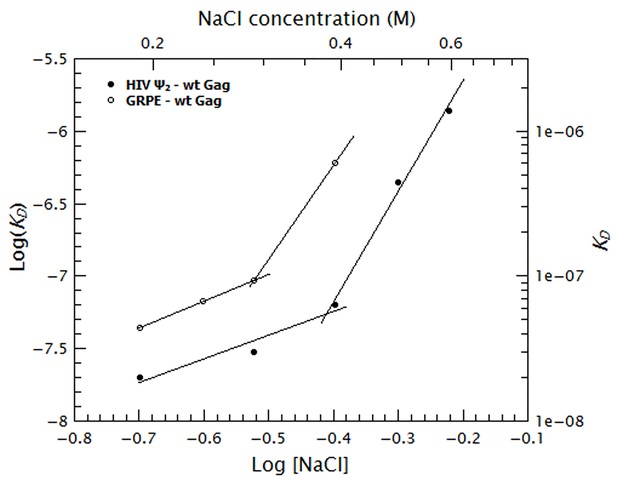
Binding of Gag to the dimeric HIV-1 Ψ has a stronger non-electrostatic component than to the GRPE RNA.
The logarithms of the KDs obtained in Figure 7 were plotted as a function of logarithm of the NaCl concentration. The fact that these plots are not linear implies that there are two different binding modes at different salt concentrations. The intercept of each fit represents the dissociation constant in the absence of electrostatic interactions (KD(1M)). At any salt concentration binding to the dimeric HIV-1 Ψ (black solid symbols) has a stronger non-electrostatic component than the GRPE RNA (black open symbols).
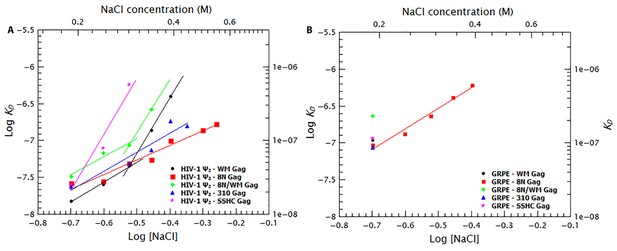
Binding to the GRPE RNA is driven by electrostatic interactions.
Binding of WM Gag (black circles), 8N Gag (red squares), 8N/WM Gag (green diamonds), 310 Gag (blue triangles) and SSHC Gag (pink stars) to the dimeric HIV-1 Ψ (A) and to the GRPE RNA (B) was measured at different ionc strengths. (A) Decreasing MA-RNA (8N), Gag-Gag (WM), MA-RNA as well as Gag-Gag (8N/WM), as well as the electrostatic interactions between NC and the RNA (310), modulated the salt dependency of the interaction with dimeric HIV-1 Ψ. (However, SSHC binding still retains a strong electrostatic component). (B) Unlike the dimeric Ψ, the affinity of these proteins for GRPE RNA, with the exception of 8N Gag (red squares), is greatly impared by increasing salt concentration; we could not detect GRPE binding to these proteins at [NaCl]≥0.25 M. The KDs calculated from Figure 9—figure supplement 1B–F and Figure 9—figure supplement 2B–F were used to generate panels A and B, respectively. A comparison of the plots of WT Gag (Figure 8) and 8N Gag (red squares in this Figure 9) is shown in Figure 9—figure supplement 3.
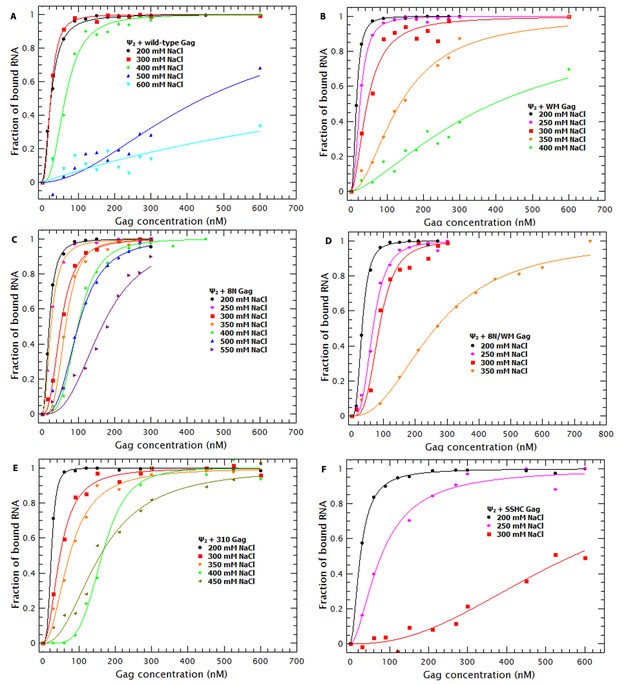
Binding plots used to generate Figures 8 and 9A.
Binding plots of the dimeric HIV-1 Ψ at different salt concentrations for different Gag proteins; (A) WT Gag, (B) WM Gag, (C) 8N Gag, (D) 8N/WM Gag, (E) 310 Gag and (F) SSHC Gag. The lines are fits of the experimental data using a cooperative model. Panel A is identical to Figure 7A.
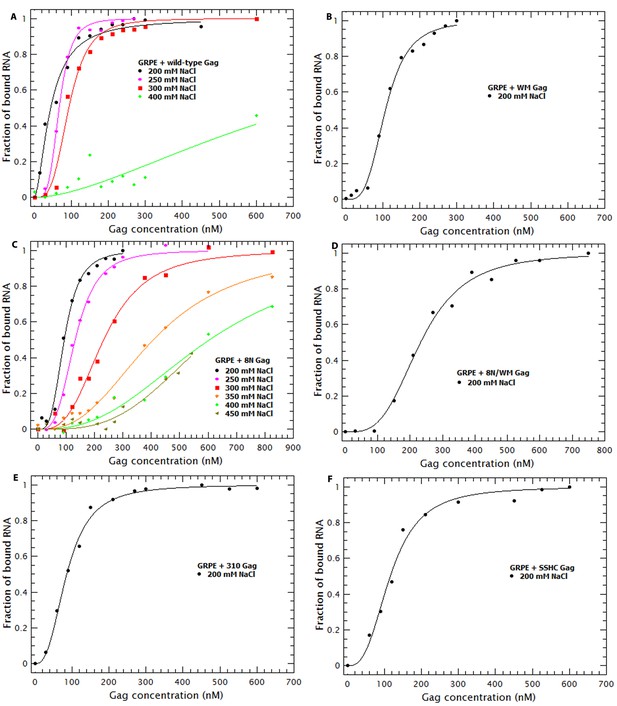
Binding plots used to generate Figures 8 and 9B.
Binding plots of GRPE RNA at different salt concentrations for different Gag proteins; (A) WT Gag, (B) WM Gag, (C) 8N Gag, (D) 8N/WM Gag, (E) 310 Gag and (F) SSHC Gag. We could not detect binding of this RNA to WM, 8N/WM, 310 and SSHC Gag at salt concentrations above 0.2 M NaCl. The lines are fits of the experimental data using a cooperative model. Panel A is identical to Figure 7B.
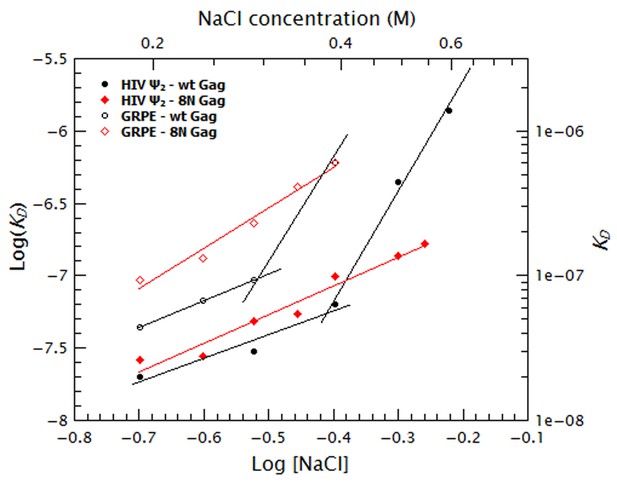
Comparison of the salt dependency of WT and 8N Gag binding to the GRPE RNA and the dimeric HIV-1 Ψ RNA.
The salt dependency for 8N Gag/Ψ (filled red diamonds) is very similar to that of the WT Gag (filled black circles), with the exception that we cannot detect binding of this protein at 0.6 M NaCl and that at 0.5–0.55M NaCl, binding to 8N Gag has a lower KD than for the WT Gag. Decreasing the charge of the MA domain increases the salt dependency for the GRPE RNA (open red diamonds).
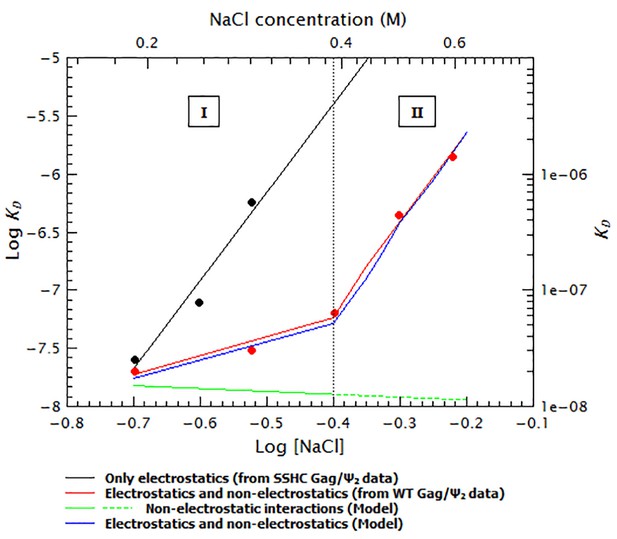
The ‘upwards bent’ salt titration indicates that binding of Gag to RNAs has a very strong non-electrostatic component.
The concave or ‘upwards bent’ shape of the salt titration data for WT Gag/Ψ2 (red circles) was obtained by using a model in which the observed KD (blue line) is the weighted sum of strong non-electrostatic (green line) and electrostatic (black line) interactions. The salt titration data for SSHC Gag/Ψ2 (black circles) was used to extract the pure electrostatic component of binding to the dimeric HIV-1 Ψ (black line) and the WT Gag/Ψ2 represents the sum of both electrostatic and non-electrostatic interactions (red line). In the first binding mode (I), binding is driven by non-electrostatic interactions (solid green line). In the second binding mode (II), a conformational change in Gag gradually decreases the relative contribution of the non-electrostatic interactions (broken green line).
Tables
Binding parameters for Gag/RNA interactions.
KD and nH values were obtained by fitting the binding curves as described in Materials and methods and in the Supplementary Information.
| WT-Gag | 8N-Gag | WM-Gag | |||||
|---|---|---|---|---|---|---|---|
| Sample | tRNA | KD (nM) | nH | KD (nM) | nH | KD (nM) | nH |
| HIV Ψ1 (monomeric) | - | 21.4 | 3.2 | 60 | 2.4 | 31.9 | 2.4 |
| HIV Ψ1 (monomeric) | + | 53.3 | 1.7 | 60 | 2.4 | 124.8 | 3.7 |
| HIV Ψ2 (dimeric) | - | 20.3 | 2.4 | 43.2 | 2.1 | 15.2 | 2.4 |
| HIV Ψ2 (dimeric) | + | 37 | 2.1 | 54.7 | 2.8 | 55.8 | 2.2 |
| MoMLV Ψ | - | 25.9 | 2.1 | 59.8 | 2.5 | 59 | 4.6 |
| MoMLV Ψ | + | 118.8 | 3.5 | 59 | 4 | ≥300 | - |
| GRPE | - | 44.2 | 1.7 | 145 | 3.9 | 111.8 | 3.8 |
| GRPE | + | 315.4 | 2.8 | 154.3 | 3.8 | >>300 | - |
Additional files
-
Supplementary file 1
Diffusion coefficients of the dye used to calibrate the FCS setup and the D of Lysozyme labeled with AF647
- https://doi.org/10.7554/eLife.27055.024





