Mitosis can drive cell cannibalism through entosis
Figures
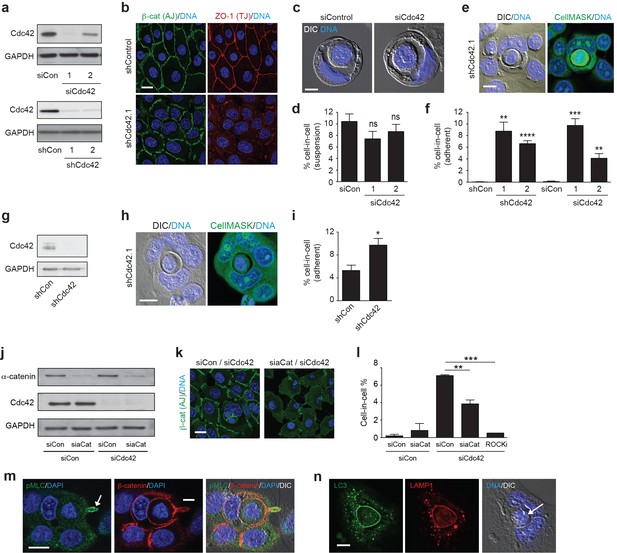
Cdc42 controls entosis in adherent epithelial cells.
(a) Control and siRNA or shRNA Cdc42-depleted 16HBE cell lysates were probed for Cdc42 and GAPDH expression by western blotting. (b) Representative confocal images of control and Cdc42-depleted 16HBE monolayers stained for β-catenin (adherens junctions), ZO-1 (tight junctions) and DNA. Scale bar = 20 μm. (c) Representative images of cell-in-cell structures formed in matrix detached control and Cdc42-depleted 16HBE cells. Cell were stained for DNA (blue) and imaged by IF/confocal and DIC. Scale bar = 5 μm. (d) Quantification of suspension cell-in-cell formation in control and Cdc42-depleted cells. >200 cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. ns = no significant difference, t-test. (e) Representative images of a cell-in-cell structure formed under adherent conditions in Cdc42-depleted 16HBE cells. Cells were stained for cell body (green) and DNA (blue) and imaged by IF/confocal and DIC. Scale bar = 10 μm. (f) Quantification of adherent cell-in-cell formation. >200 16HBE cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. **p<0.002; ***p<0.0002; ****p<0.0001, t-test. (g) Control and shRNA Cdc42-depleted MCF7 cell lysates were probed for Cdc42 and GAPDH expression by western blotting. (h) Representative images of a cell-in-cell structure formed under adherent conditions in Cdc42-depleted MCF7 cells. Cells were stained for cell body (green) and DNA (blue) and imaged by IF/confocal and DIC. Scale bar = 10 μm. (i) Quantification of adherent cell-in-cell formation. >200 MCF7 cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. *p<0.02, t-test. (j) Lysates from 16HBE cells co-depleted of Cdc42 and α-catenin (aCat) were probed for α-catenin, Cdc42 and GAPDH by western blotting. (k) Representative confocal images of 16HBE cells co-depleted of Cdc42 and siControl or α-catenin and stained for β-catenin (green) and DNA (blue). Scale bar = 20 μm. (l) Quantification of cell-in-cell structures in adherent 16HBE cells treated with siCdc42 and siControl or siα-catenin, treated −/+10 μM Y-27632 (ROCKi), for 3 days. >200 cells were scored per sample/experiment, across three separate experiments. Error bars denote mean±SEM. **p<0.002; ***p<0.0002, t-test. (m) Confocal images of a forming cell-in-cell structure in adherent, Cdc42-depleted 16HBE cells fixed and costained for pMLC2 (S19; green), β-catenin (red) and DNA (Hoechst, blue). The arrowhead indicates the tail of the internalising cell. Scale bar = 10 μm. (n) Cell-in-cell structures in Cdc42-depleted 16HBE cells were fixed and costained for LC3 (green), LAMP1 (red) and DNA (blue), and imaged by IF/confocal and DIC. The arrowhead indicates a dying internalised cell. Scale bar = 10 μm.
-
Figure 1—source data 1
- https://doi.org/10.7554/eLife.27134.004
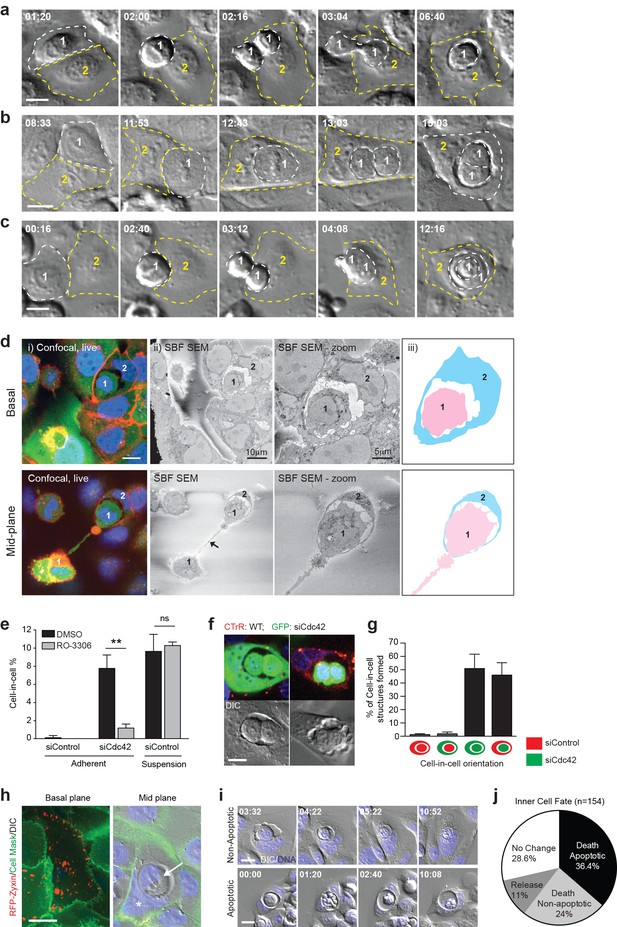
Mitosis drives entotic cell cannibalism in adherent cells.
(a–c) Cdc42-depleted 16HBE cells were analysed by timelapse microscopy. Three different configurations of cell-in-cell formation are shown with timestamps (hr:min). In each case, a mitotic cell (Cell 1, outlined white) enters an adherent entotic host (Cell 2, outlined yellow). Scale bars = 10 μm. (d) Cdc42-depleted 16HBE cells were analysed by 3D-CLEM. (i) Live confocal sections from basal and mid-planes of a forming cell-in-cell structure, stained for plasma membrane (red), cell body (green) and DNA (blue). A mitotic daughter (Cell 1) is shown internalising into an adherent neighbour (Cell 2). Scale bar = 10 μm. (ii) Corresponding serial blockface scanning electron microscopy (SBF-SEM) images of the same forming structure. The arrowhead marks the midbody between daughter cells. (iii) Cartoon outline of cell-in-cell structure from (ii). (e) Quantification of cell-in-cell formation among control and Cdc42-depleted 16HBE cells, under adherent or suspension conditions, in the presence or absence of RO-3306 (5 μM), a Cdk1 inhibitor that induces G2/M arrest. >200 cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. **p<0.002; ns = no significant difference, t-test. (f) Representative confocal and DIC images of adherent cell-in-cell structures in wild-type-16HBE (red) and Cdc42-depleted GFP-16HBE (green) co-cultures. Scale bar = 15 μm. (g) Quantification of cell-in-cell formation between WT and Cdc42-depleted 16HBE co-cultures as described in (f). >50 cell-in-cell structures were imaged per condition/experiment, across three separate experiments. Error bars denote mean±SEM. (h) Cdc42-depleted 16HBE cells were mixed with wild-type cells expressing RFP-zyxin, incubated for 3 days then stained for plasma membrane (green) and DNA (blue) and imaged by live confocal and DIC microscopy. A representative adherent cell-in-cell structure is shown, with basal and mid-plane sections presented; asterix = host cell nucleus, arrowhead = internalised cell. Scale bar = 15 μm. (i) Inner cell fate was analysed in Cdc42-depleted adherent 16HBE entotic structures, stained for DNA (blue). Representative timelapse series show non-apoptotic and apoptotic inner cell death; timestamps = (hr:min). Scale bar = 10 μm. (j) Quantification of inner cell fate over 20 hr in adherent Cdc42-depleted 16HBE structures. 154 cell-in-cell structures were analysed over three independent experiments.
-
Figure 2—source data 1
- https://doi.org/10.7554/eLife.27134.006
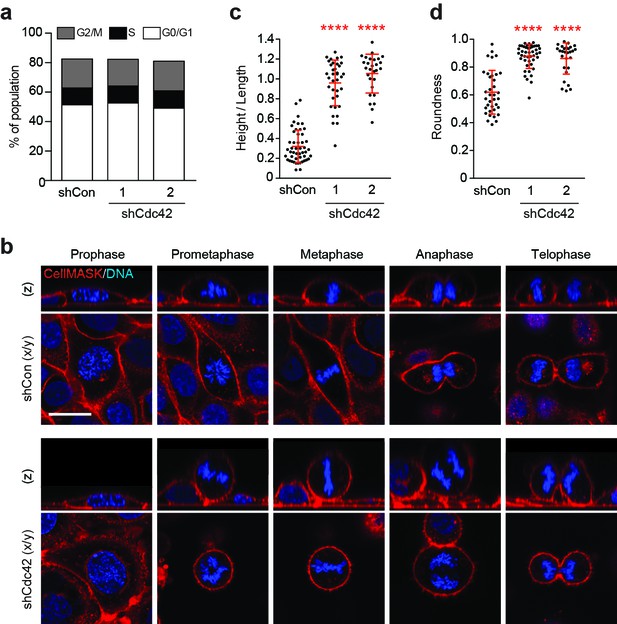
Cdc42 controls mitotic deadhesion and rounding in polarised epithelial cells.
(a) Control and Cdc42-depleted 16HBE cells were fixed and stained with propidium iodide. DNA content was analysed by FACS. (b) Control and Cdc42-depleted 16HBE cells were stained for plasma membrane (red) and DNA (blue), and analysed by live confocal microscopy. Representative sections and z-stacks of different phases of mitosis are shown. Scale bar = 20 μm. Quantification of (c) mitotic spreading (cell height/length) and (d) mitotic rounding (where 1 = a perfect circle) in control and Cdc42-depleted cells. >10 metaphase cells were imaged per sample/experiment, across three independent experiments. Error bars denote mean±SD. ****p<0.0001, Mann-Whitney U test.
-
Figure 3—source data 1
- https://doi.org/10.7554/eLife.27134.013
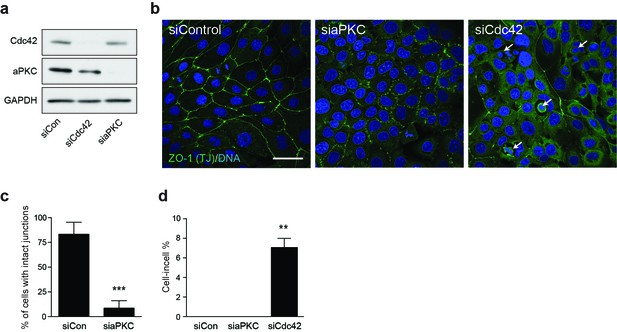
Cdc42 controls adherent cell-in-cell formation, but aPKC does not.
(a) Lysates from control and siCdc42 and siaPKC 16HBE cells were probed for Cdc42, aPKC and GAPDH by western blot. (b) Representative confocal images of siRNA-treated cells stained for ZO-1(green), to visualise tight junctions, and DNA (blue). Arrows indicate cell-in-cell structures. Scale bar = 50 mm. Quantification of (c) junctions and (d) cell-in-cell formation in siRNA treated cells. **p<0.002; ***p<0.0008, t-test. Depletion of aPKC disrupts tight junction formation, like Cdc42, but does not induce entosis.
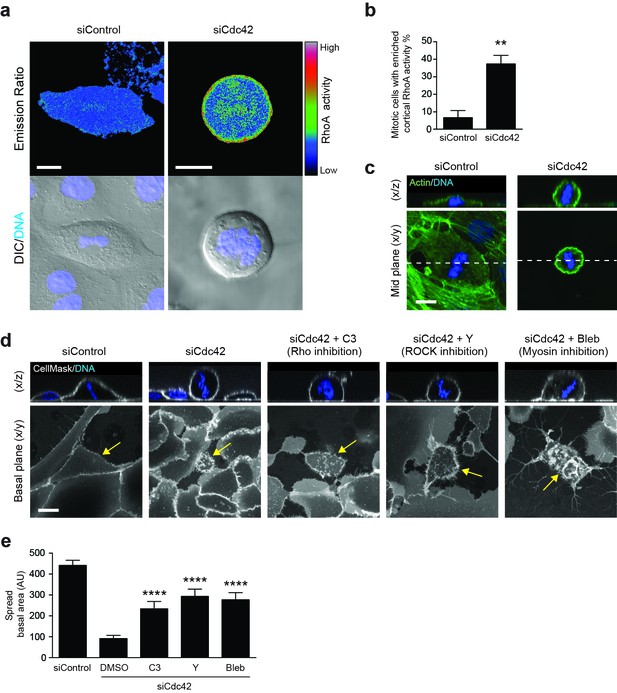
RhoA activity is spatiotemporally regulated by Cdc42 and controls mitotic spreading.
(a) 16HBE cells expressing a RhoA FRET biosensor were treated with siControl or siCdc42. Three days later, cells were subjected to live confocal imaging for CFP (FRET donor), YFP (FRET acceptor), DNA and DIC. RhoA activity is represented by the FRET/CFP emission ratio. Scale bars = 10 μm. (b) RhoA activity was measured in >30 metaphase cells per condition, across four independent experiments, and cortical enrichment of active RhoA was scored. **p<0.003, t-test. (c) Control or Cdc42-depleted 16HBE cells were fixed and stained for actin (green) or DNA (blue), and metaphase cells were imaged by IF/confocal. Representative sections and z-stacks are shown. Scale bar = 10 μm. (d) 16HBE cells were treated with siControl or siCdc42 and incubated for 3 days. Cdc42-depleted cells were then incubated with C3 (Rho inhibitor; 1 μg/ml), Y-27632 (ROCK inhibitor; 10 μM) or Blebbistatin (myosin inhibitor; 100 μM) for a further 4 hr. Live cells were stained for plasma membrane (white) and DNA (blue) and imaged by IF/confocal to assess metaphase morphology; scale bar = 15 μm. Representative z-stacks and basal sections are shown. (e) The spread basal area of each metaphase cell was measured. >15 cells were scored/condition, across three independent experiments. ****p<0.0001.
-
Figure 4—source data 1
- https://doi.org/10.7554/eLife.27134.016
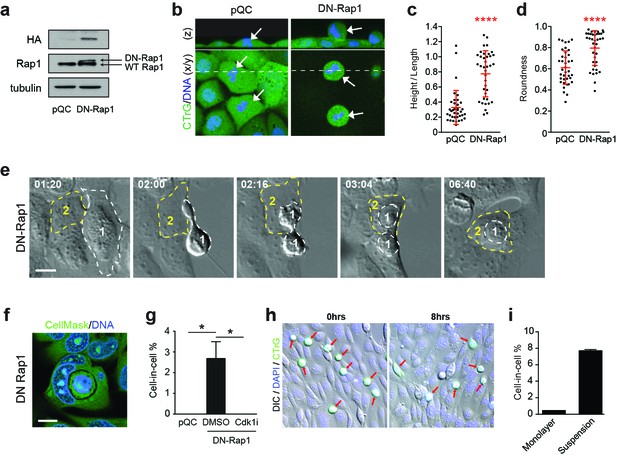
Enhanced mitotic deadhesion and rounding can drive entosis.
(a) Control (pQC) and DN-Rap1-HA expressing 16HBE cells were probed for HA, Rap1 and tubulin by western blot. (b) Control and DN-Rap1 expressing 16HBE cells were stained for cell body (green) and DNA (blue) and analysed by live confocal microscopy. Representative midplane x/y, and z sections through the dashed line, are presented. Arrowheads indicate metaphase cells, as identified by nuclear morphology. Quantification of (c) mitotic spreading (cell height/length) and (d) mitotic rounding (where 1 = a perfect circle) in control and DN-Rap1 16HBE cells. >10 metaphase cells were imaged per sample/experiment, across three independent experiments. Error bars denote mean±SD. ****p<0.0001, Mann-Whitney U test. (e) DN-Rap1 expressing 16HBE cells were analysed by timelapse microscopy. A mitotic cell (Cell 1) is outlined in white, the adherent entotic host (Cell 2) in yellow. Timestamps are indicated (hr:min) and scale bar = 10 μm. (f) Representative confocal image of an adherent cell-in-cell structure in DN-Rap1 expressing 16HBE cells, fixed and stained for the cell body (green) and DNA (blue). Scale bar = 10 μm. (g) Quantification of cell-in-cell formation in adherent control and DN-Rap1 cells, treated −/+ a Cdk1 inhibitor that induces G2/M arrest (5 μM RO-3306; Cdk1i). >250 cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. *p<0.03, t-test. (h) Representative images from a co-culture of suspension wild type 16HBE cells, labelled with CellTracker (green), overlaid on an existing monolayer of wild type 16HBE cells stained for DNA (blue). Co-cultures were monitored for 8 hr by timelapse microscopy. DIC/IF images are shown at time 0 and 8 hr; the detached population (green) are highlighted with red arrows; these cells can persist throughout the timecourse and very rarely penetrate adherent hosts. (i) Quantification of cell-in-cell formation under adherent conditions as described in (h), and under suspension conditions. >300 cells were counted per sample/experiment, across three independent experiments. Error bars denote mean±SEM.
-
Figure 5—source data 1
- https://doi.org/10.7554/eLife.27134.018
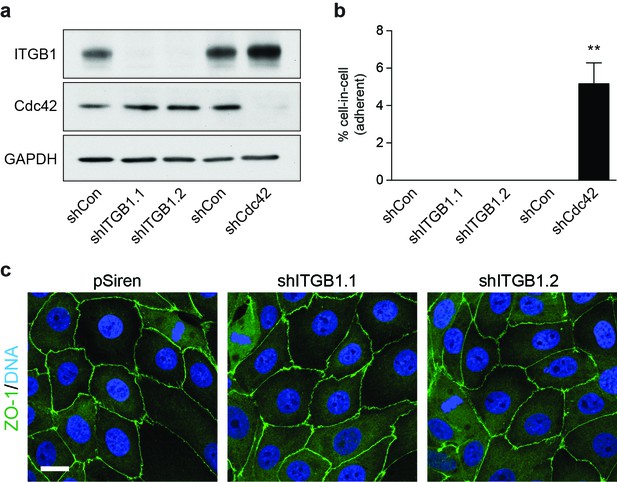
Loss of β1 integrin (ITGB1) does not induce adherent cell-in-cell formation.
(a) Lysates from control and shITGB1 and shCdc42 16HBE cells were probed for ITGB1, Cdc42 and GAPDH by western blot. (b) Quantification of cell-in-cell formation in shRNA-treated cells. >300 cells were counted per sample/experiment, across three independent experiments. Error bars denote mean±SEM. **p<0.002; t-test. (c) Representative confocal images of shITGB1-treated cells stained for ZO-1 (green), to visualise tight junctions, and DNA (blue). Note, no cell-in-cell structures are observed and mitotic cells remain spread in ITGB1-depleted cells. Scale bar = 15 μm.
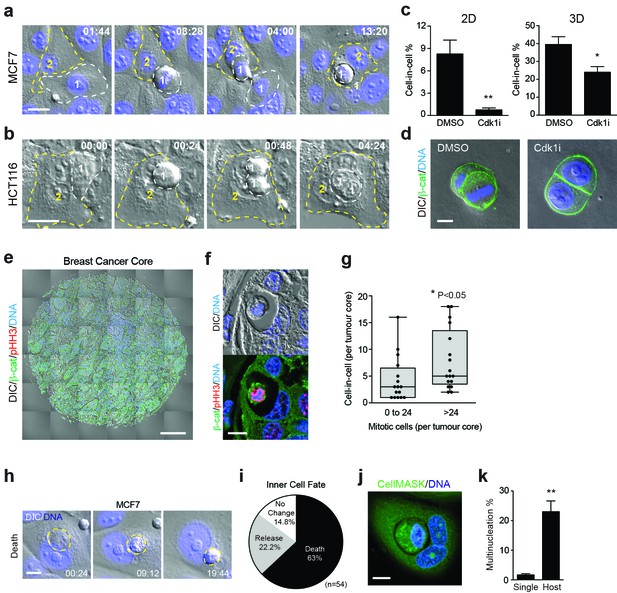
Mitosis-induced entosis occurs in cancer cell lines and human tumours, with pleiotropic effects.
(a–b) Adherent cancer cell lines MCF7 (breast) and HCT116 (colorectal) were analysed by timelapse microscopy; representative cell-in-cell formation events are shown. In each case, a mitotic cell (Cell 1, outlined in white) is internalised by an adherent neighbour (Cell 2, outlined in yellow). Timestamps are shown (hr:min) and scale bar = 20 μm. (c) Quantification of cell-in-cell formation in adherent MCF7 cells cultured in 2D or 3D, treated +/− a Cdk1 inhibitor that induces G2/M arrest (5 μM RO-3306; Cdk1i). >300 cells for 2D and >50 cells for 3D were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. **p<0.002; *p<0.02, t-test. (d) MCF7 cells seeded in 3D matrigel were treated −/+5 μM RO-3306 overnight, then fixed and stained for β-catenin (green) and DNA (blue). Two to four cell cysts were imaged to assess mitotic status and cell-in-cell formation. Representative sections are shown; scale bar = 10 μm. (e) Human breast invasive ductal carcinoma. A tumour microarray was stained for β-catenin (green), pS10-Histone H3 (red, mitotic marker) and DNA (blue) and imaged in full by DIC and IF/confocal. A tiled confocal image is presented for one core. Scale bar = 200 μm. (f) Entosis in a human invasive breast ductal carcinoma. A representative cell-in-cell structure is shown by DIC and IF/confocal; notably the inner cell is mitotic as judged by pHH3. Scale bar = 10 μm. (g) Quantification of mitotic index and cell-in-cell formation among human breast invasive ductal carcinomas. The median number of mitotic cells/core is 24. *p<0.05, Mann-Whitney test. (h) Representative timelapse series showing inner cell death in an MCF7 cell-in-cell structure, stained for DNA (blue). The internalised cell is outlined in yellow; its corpse shrinks over time. Timestamps are indicated (hr:min) and scale bar = 10 μm. (i) Quantification of inner cell fate in MCF7 entotic structures over 20 hr. Fifty-four cell-in-cell structures were analysed over three independent experiments. (j) Representative image of a multinucleated, entotic host cell in adherent MCF7 stained for cell body (green) and DNA (blue). Scale bar = 10 μm. (k) Quantification of MCF7 multinucleation in single cells versus entotic hosts cells. Error bars denote mean±SEM across three independent experiments. **p<0.008, t-test.
-
Figure 6—source data 1
- https://doi.org/10.7554/eLife.27134.022
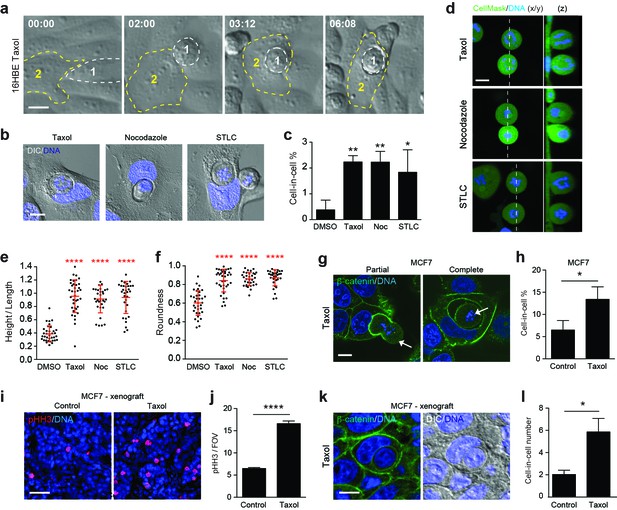
Paclitaxel/taxol treatment promotes mitotic deadhesion, rounding and entosis.
(a) Representative timelapse images of adherent 16HBE cells treated with 1 μM taxol. Cell 1 (outlined white) rounds up in prometaphase and subsequently penetrates an adherent interphase neighbour (Cell 2, outlined yellow). Timestamps are shown (hr:min) and scale bar = 10 μm. (b) Representative confocal/DIC images of adherent cell-in-cell structures in 16HBE treated with taxol (1 μM), nocodazole (100 ng/ml) or STLC (20 μM) for 24 hr. Cells were stained for DNA (blue), scale bar = 10 μm. (c) Quantification of drug-induced cell-in-cell formation. >150 cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. **p<0.002; *p<0.02, t-test. (d) Mitotic morphology of 16HBE cells treated with taxol (1 μM), nocodazole (100 ng/ml) or STLC (20 μM) for 24 hr. Live cells were stained for cell body (green) and DNA (blue). Midplane x/y, and z sections through the dashed line, are presented. Scale bar = 10 μm. Quantification of (e) mitotic spreading (cell height/length) and (f) mitotic rounding (where 1 = a perfect circle) in control and drug-treated cells. >15 cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SD. ****p<0.0001, Mann-Whitney U test. (g) Representative confocal images of partially and completely formed cell-in-cell structures in MCF7 cells treated with taxol (1 μM), and stained for β-catenin (green) and DNA (blue). The arrowheads point to prometaphase arrested cells internalised by adherent, interphase neighbours. Scale bar = 10 μm. (h) Quantification of taxol-induced entosis in MCF7. >150 cells were counted per sample/experiment, across three separate experiments. Error bars denote mean±SEM. *p<0.04, t- test. (i) Confocal images of MCF7 xenografts treated −/+ taxol for 24 hr and stained for phospho-Histone H3 (pHH3, red) and DNA (blue). Scale bar = 50 μm. (j) Quantification of pHH3-positive, mitotic cells in MCF7 mouse xenografts treated −/+ taxol for 24 hr. ****p<0.0001, t-test. (k) Representative confocal and DIC images of an entotic cell-in-cell structure in a taxol-treated MCF7 xenograft, stained for β-catenin (green) and DNA (blue). Scale bar = 10 μm. (l) Quantification of cell-in-cell formation in MCF7 mouse xenografts treated −/+ taxol for 24 hr. *p<0.01, t-test.
-
Figure 7—source data 1
- https://doi.org/10.7554/eLife.27134.026
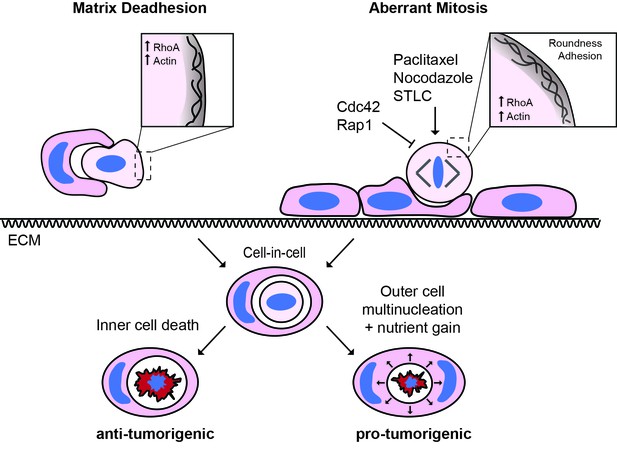
The triggers and consequences of entosis in cancer.
Entosis can be triggered among epithelial cells through either matrix deadhesion or aberrant mitosis. Mitotic entosis is associated with enhanced deadhesion and rounding during cell division, which can be induced by inhibition of Cdc42 or Rap1, or through prometaphase arrest. RhoA activity is important in both suspension and mitosis-induced entosis, driving ROCK-dependent myosin activation. Regardless of the triggering mechanism, entosis promotes both inner cell death and outer cell nutrient gain and multi-nucleation, with the potential to confer both anti- and pro-tumorigenic effects.
Videos
Mitosis-driven entosis in adherent Cdc42-depleted 16HBE cells.
DIC images from Widefield timelapse. Cell 1 engulfed by cell 2 post mitosis.
Mitosis-driven entosis in adherent Cdc42-depleted 16HBE cells.
DIC images from Widefield timelapse. Cell 1 is engulfed by cell 2 during mitosis.
Mitosis-driven entosis in adherent Cdc42-depleted 16HBE cells.
DIC images from Widefield timelapse. Cell 1 daughters engulf each other and are then engulfed by cell 2.
Live cell confocal z-stack of a forming cell-in-cell structure in Cdc42-depleted adherent 16HBE cells.
Cells are stained with CellTracker green (cell body), CellMASK, red (membrane) and Hoechst (DNA, blue).
Corresponding Serial Block Face SEM z-stack of forming cell-in-cell structure in Video 4.
https://doi.org/10.7554/eLife.27134.011Mitosis-driven entosis in adherent 16HBE cells expressing DN-Rap1.
DIC images from Widefield timelapse. A daughter of cell 1 is engulfed by cell 2.
Mitosis-driven entosis in adherent MCF7 cells.
DIC and DNA(Hoechst) images from Widefield timelapse. A daughter of cell 1 is engulfed by cell 2.
Mitosis-driven entosis in adherent HCT116 cells.
DIC images from Widefield timelapse. Cell 1 is engulfed by cell 2 during mitosis.
Mitosis-driven entosis in adherent 16HBE cells treated with taxol (1 μM).
DIC images from Widefield timelapse. Cell 2 enters mitosis and is engulfed by cell 1.
Additional files
-
Supplementary file 1
Human breast invasive ductal carcinoma tissue microarray.
Accompanying tumour information for the Biomax tumour microarray BR1505b used in Figure 6d.
- https://doi.org/10.7554/eLife.27134.029





