Genetic variation in adaptability and pleiotropy in budding yeast
Figures
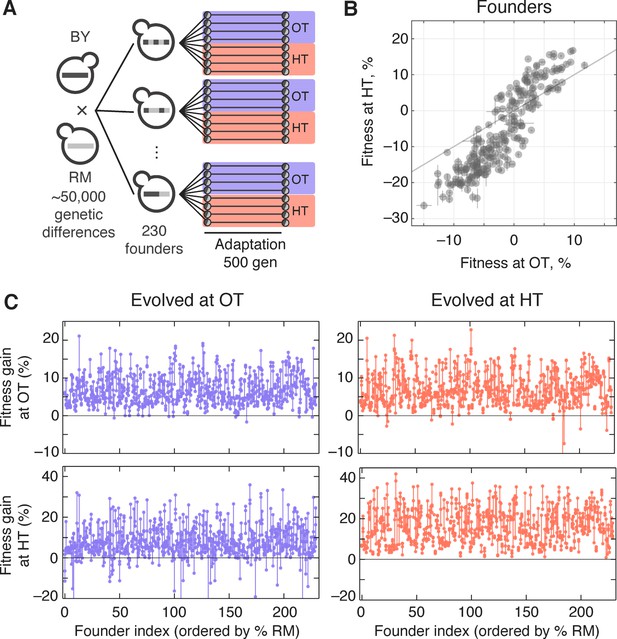
Experimental design and fitness gains in evolved populations.
(A) Schematic of the experimental design. (B) Fitness of all 230 founders at both OT and HT. (C) Fitness of all evolved lines in both OT and HT environments, after 500 generations of adaptation in one of the environments. Each point represents the fitness of one population; vertical lines connect populations descended from the same founder. Founders are ordered by the fraction of their genome derived from the RM parent.
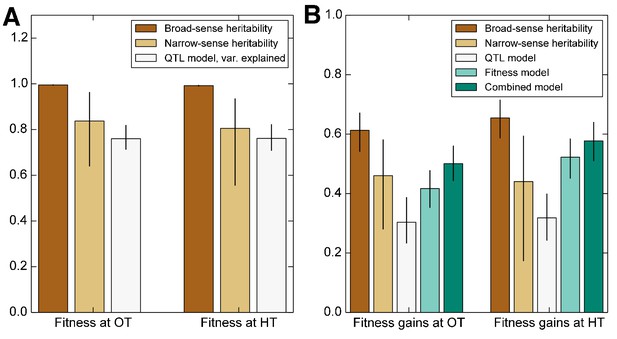
Heritability of fitness and adaptability.
Tan bars denote broad-sense and narrow sense heritability of (A) founder fitness and (B) fitness gain after evolution in the indicated environment (i.e. adaptability). White bars denote the heritability explained by a linear model based on the QTLs identified as affecting initial fitness or adaptability (Materials and methods). Green bars indicate the heritability of adaptability explained by initial fitness in that condition. Error bars indicate 95% confidence intervals.
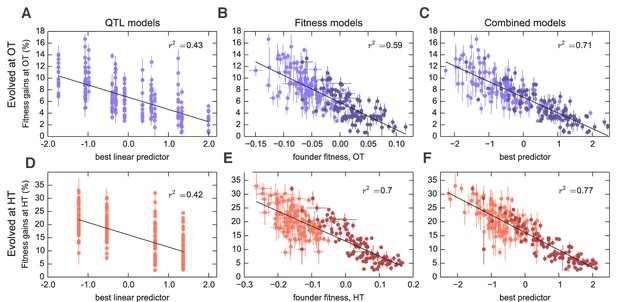
Models to predict adaptability.
Each point represents the average fitness change of four populations descended from the same founder after 500 generations of evolution in the (A–C) OT environment or (D–F) HT environment. Error bars denote s.e.m. Lines show the predictions of different models; indicates the fraction of the variance explained, which is similar to the fraction of the broad-sense heritability explained by the model. (A,D) Linear QTL model, where the -axis is the linear combination of identified QTLs that best predicts the fitness gains in each environment. (B,E) Model where founder fitness is used as predictor for fitness gain. (C,F) Combined model, where the -axis is the linear combination of initial fitness and the identified QTLs that best predicts the fitness gains in each environment. Dark and light shades represent BY and RM alleles at the KRE33 locus respectively. For model parameters, see Figure 3—source data 1.
-
Figure 3—source data 1
Parameters for combined fitness and QTL models for adaptability.
- https://doi.org/10.7554/eLife.27167.006
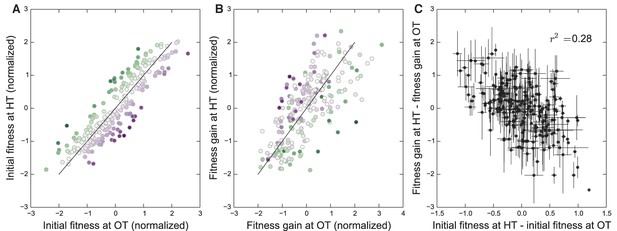
Initial founder fitness and adaptability in both environments.
(A) Normalized fitness of founders at OT and HT (Materials and methods). Each point is colored according to its deviation from the overall correlation in initial fitnesses. Error bars are omitted for clarity. (B) Normalized fitness increments for all founders (Materials and methods). Colors are the same as in (A); note that points above the diagonal in (A) tend to be found below the diagonal in (B), and vice versa. (C) Difference in fitness increments after adaptation in each environment as a function of the difference in normalized initial founder fitness in HT versus OT. Error bars denote s.e.m.
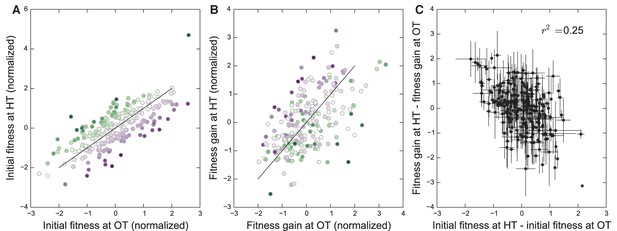
The effect of fitness in the home environment on adaptability, controlling for the effect of the KRE33 allele.
Same as Figure 4, except fitnesses and fitness increments are normalized for the two founder groups with different KRE33 alleles separately.
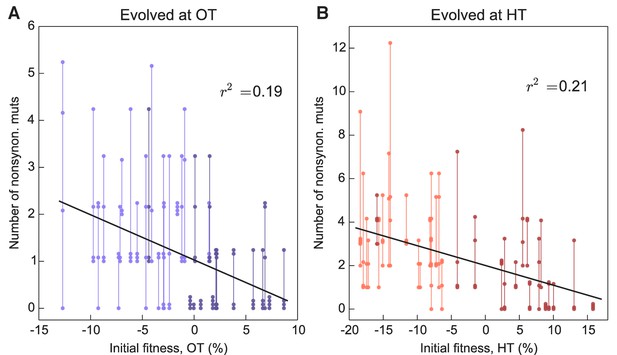
Number of fixed nonsynonymous mutations.
Each point represents the number of fixed nonsynonymous mutations in a population evolved at (A) OT or (B) HT. Populations descended from the same founder are connected by vertical lines for visual convenience, with founders ordered according to their fitness in the corresponding environment. Dark and light shades represent BY and RM alleles at the KRE33 locus respectively.
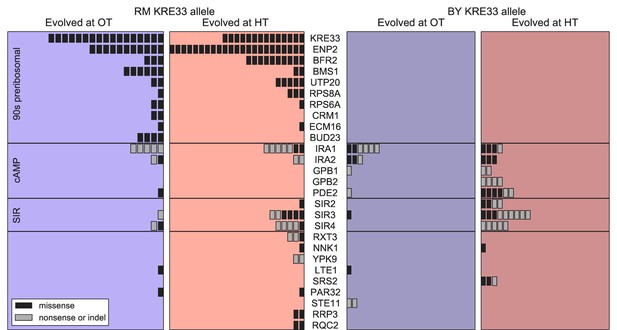
Distribution of mutations in multi-hit genes across evolved populations.
The number of sequenced populations with a de novo mutation in each multi-hit gene, for each environment and founder KRE33 identity. Genes are organized by functional groups. For mutual information metrics, see Figure 6—source data 1.
-
Figure 6—source data 1
Mutual information between de novo mutations and the KRE33 allele, the evolution environment, and the genotype.
- https://doi.org/10.7554/eLife.27167.011
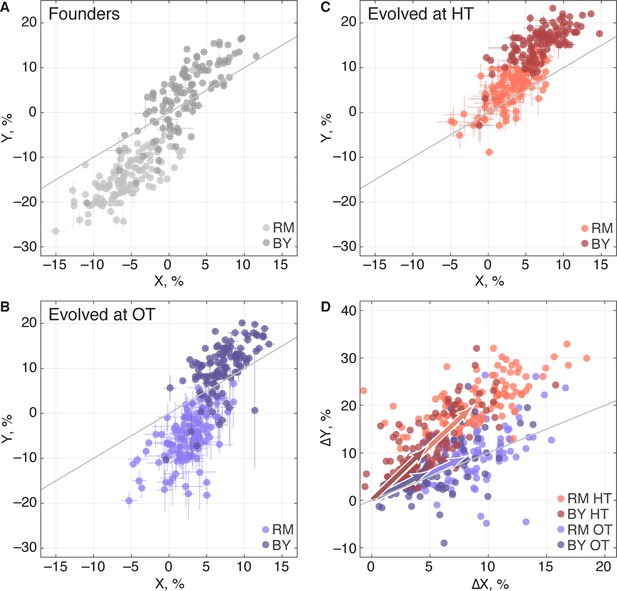
Pleiotropic consequences of adaptation.
(A) The fitness of each founder in the OT environment () and HT environment (). Error bars represent s.e.m. over technical replicates. Dark and light points represent strains carrying the BY and RM alleles at the KRE33 locus respectively. (B,C) Average fitness of all populations descended from each founder. Error bars represent s.e.m. over replicate populations. Orange and blue colors represent strains evolved at HT and OT, respectively; dark and light shades represent BY and RM alleles at the KRE33 locus respectively. (D) Average fitness increment of all populations descended from each founder; error bars omitted for clarity. Arrows point to the centroids of the respective point clouds.
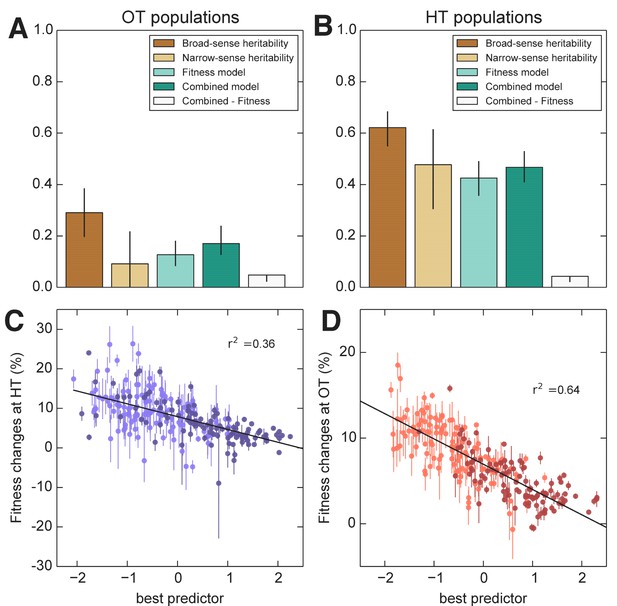
Heritability and predictability of pleiotropic fitness gains.
(A) Heritability of the fitness gains in the HT environment after evolution at OT, , and the variance explained by fitness and combined fitness/QTL models. The final (grey) bar indicates the median difference in variance explained between the combined fitness/QTL model and the fitness model under jackknife resampling (Materials and methods). (B) Heritability of and the variance explained by fitness and combined fitness/QTL models. The final (grey) bar is as in A. (C) Predicted and actual values of based on initial fitness at HT and QTL loci (Supplementary file 5, column 9). Each point shows the mean over populations descended from the same founder. Error bars denote s.e.m. over technical replicates (-axis) or over replicate evolved populations (-axis). Dark and light points represent strains carrying the BY and RM alleles at the KRE33 locus respectively. (D) Predicted and actual values of based on initial fitness at OT and QTL loci (Supplementary file 5, column 10). For model parameters, see Figure 8—source data 1.
-
Figure 8—source data 1
Parameters for combined fitness and QTL models for pleiotropy.
- https://doi.org/10.7554/eLife.27167.015
Tables
Heritability of fitness, adaptability, and pleiotropy, along with the fraction of variance explained by QTL, fitness, and combined models.
Row 5 represents the median added variance explained by the fitness model over the QTL model under a jacknife over segregants (Materials and methods); Row 7 similarly represents the added variance explained by the combined model over the fitness model. Paired numbers in parentheses denote 95% confidence intervals; single numbers in parentheses denote lower bounds (5th percentile) on added variances.
| Fitnessat OT | Fitnessat HT | Adaptabilityat OT | Adaptabilityat HT | PleiotropyHT pops at OT | PleiotropyOT pops at HT | |
|---|---|---|---|---|---|---|
| H2 | 0.995 (0.992, 0.997) | 0.992 (0.988, 0.994) | 0.613 (0.541, 0.672) | 0.654 (0.586, 0.716) | 0.622 (0.552, 0.680) | 0.291 (0.193, 0.394) |
| 0.837 (0.639, 0.964) | 0.805 (0.555, 0.936) | 0.460 (0.280, 0.582) | 0.440 (0.173, 0.595) | 0.477 (0.308, 0.636) | 0.091 (0, 0.228) | |
| , QTL models | 0.760 (0.712, 0.820) | 0.761 (0.707, 0.823) | 0.303 (0.232, 0.388) | 0.318 (0.242, 0.340) | NA | NA |
| , founder fitness | NA | NA | 0.417 (0.352, 0.478) | 0.522 (0.451, 0.585) | 0.425 (0.361, 0.492) | 0.127 (0.082, 0.182) |
| Added variance fitness vs. QTLs | NA | NA | 0.105 (0.047) | 0.199 (0.143) | NA | NA |
| , combined models | NA | NA | 0.501(0.442, 0.561) | 0.577 (0.510, 0.641) | 0.467 (0.411, 0.534) | 0.171 (0.126, 0.235) |
| Added variance combined vs. fitness | NA | NA | 0.087 (0.060) | 0.057 (0.035) | 0.047 (0.021) | 0.044 (0.022) |
Average fitness increments of populations in home and away environments, in percent.
Numbers in parentheses denote 95% confidence intervals.
| Kre33 -RM | Kre33 -BY | |||
|---|---|---|---|---|
| Evolved at OT | Evolved at HT | Evolved at OT | Evolved at HT | |
| Fitnessat OT, % | 8.33(8.27, 8.38) | 8.86(8.81, 8.92) | 5.01(4.94, 5.07) | 4.834.76, 4.90 |
| Fitnessat HT, % | 9.57(9.42, 9.77) | 20.33(20.18, 20.53) | 6.25(6.12, 6.38) | 11.51(11.38, 11.64) |
Additional files
-
Supplementary file 1
Genotypes of segregants used in this study.
Note this data is adapted from Bloom et al. (2013), with marker loci that are redundant for this subset of the segregant panel removed.
- https://doi.org/10.7554/eLife.27167.016
-
Supplementary file 2
Fitness measurements in the OT and HT environments for each founder and descendant population.
- https://doi.org/10.7554/eLife.27167.017
-
Supplementary file 3
All mutations called at frequencies of in evolved populations.
- https://doi.org/10.7554/eLife.27167.018
-
Supplementary file 4
Annotated sequence for pEJ03-mCitrine-KanMX-HO.
- https://doi.org/10.7554/eLife.27167.019
-
Supplementary file 5
QTL locations, and genes within confidence intervals, for all traits.
- https://doi.org/10.7554/eLife.27167.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.27167.021





