Loss of the melanocortin-4 receptor in mice causes dilated cardiomyopathy
Figures
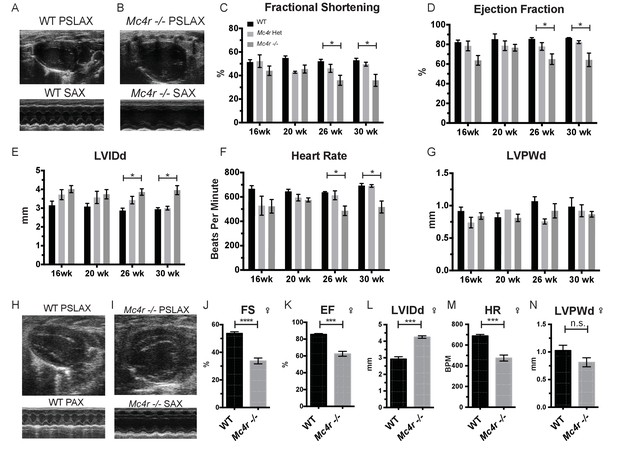
Mc4r deletion causes progressive cardiac dilatation and reduced contractility.
(A) Representative B-mode Parasternal long axis (PSLAX) image at diastole (top) and M-mode short axis (SAX) image of male WT C57B6/J mouse at 26 weeks of age. (B) Representative B-mode PSLAX image at diastole (top) and M-mode SAX image of male Mc4r−/− mouse 26 weeks of age. (C) Fractional Shortening (FS) of male WT, Mc4r−/+, and Mc4r−/− mice at 16, 20, 26 and 30 weeks of age. (2-way ANOVA Sidak multiple comparison test, p<0.05, n = 5–6) (D) Ejection Fraction (2-way ANOVA Sidak multiple comparison test, p<0.05, n = 5–6) (E) Left ventricular internal dimension during diastole (LVIDd) (2-way ANOVA Sidak multiple comparison test, p<0.05, n = 5–6) (F) Heart Rate (2-way ANOVA Sidak multiple comparison test, p<0.05, n = 5–6) (G) Left ventricle posterior wall thickness during diastole (LVPWd) (2-way ANOVA Sidak multiple comparison test, p>0.05, n = 5–6) (H) Representative B-mode PSLAX image at diastole (top) and M-mode SAX image of female WT C57B6/J mouse at 26 weeks of age. (I) Representative B-mode PSLAX image at diastole (top) and M-mode SAX image of female Mc4r−/− C57B6/J mouse 26 weeks of age. (J) Fractional shortening of female WT and Mc4r−/− mice at 26 weeks. (Student's t-test p<0.0001, n = 5) (K) Ejection fraction of female WT and Mc4r−/− mice at 26 weeks. (Student's t-test p<0.001, n = 5) (L) LVIDd (Student's t-test p<0.0001, n = 5) (M) HR (Student's t-test p<0.001, n = 5–6) (N) LVPWd (Student's t-test p=0.1185, n = 5 ).
-
Figure 1—source data 1
Source data for Figure 1C–G and J-N.
This source file provides mean, SD, sample size and statistical test information for all echocardiography analyses in Figure 1, of which examples are shown in Figure 1A–B and H-I.
- https://doi.org/10.7554/eLife.28118.004
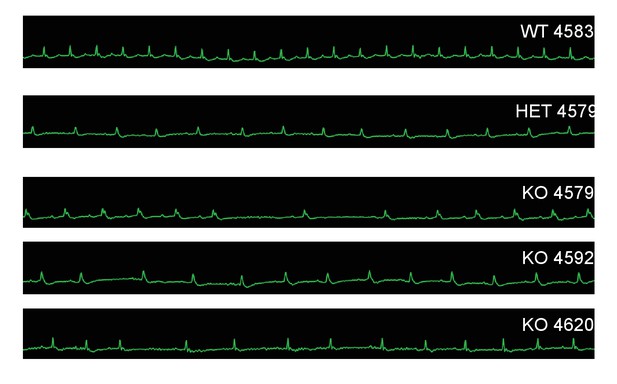
Mc4r−/− animals display bradycardic arrhythmias.
Rhythm strips of 30 week old WT, Mc4r-/+ and Mc4r−/− animals taken during echocardiograms. Mc4r−/− mice display sinus bradycardia with abnormal rhythmicity.
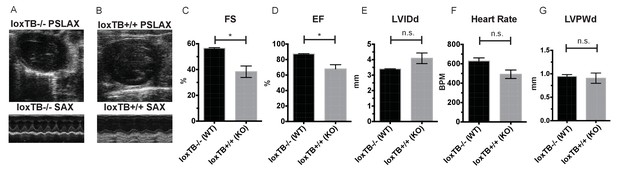
Mc4r loxTB animals phenocopy the reduced cardiac function seen in Mc4r−/− mice.
(A) Representative B-mode PSLAX image at diastole (top) and M-mode SAX image of male Mc4r loxTB -/- (WT) mouse at 35 weeks of age. (B) Representative B- mode PSLAX image at diastole (top) and M-mode SAX image of male Mc4r loxTB +/+ (Mc4r−/−) mouse 35 weeks of age. (C) Fractional Shortening (FS) of male Mc4r loxTB -/- and Mc4r loxTB +/+mice at 35 weeks (Student’s t-test, p<0.05, n = 5) (D) Ejection Fraction (Student’s t-test, p<0.05, n = 5). (E) Left ventricular internal dimension; diastole (LVIDd) (Student’s t-test, p=0.0838, n = 5). (F) Heart Rate (Student’s t-test, p>0.05, n = 5). (G) Left ventricle posterior wall thickness; diastole (Student’s t-test, p<0.05, n = 5).
-
Figure 3—source data 1
Source data for Figure 3C–G.
This source file provides mean, SD, sample size and statistical test information for all echocardiography analyses in Figure 3, of which examples are shown in Figure 3A–B.
- https://doi.org/10.7554/eLife.28118.007
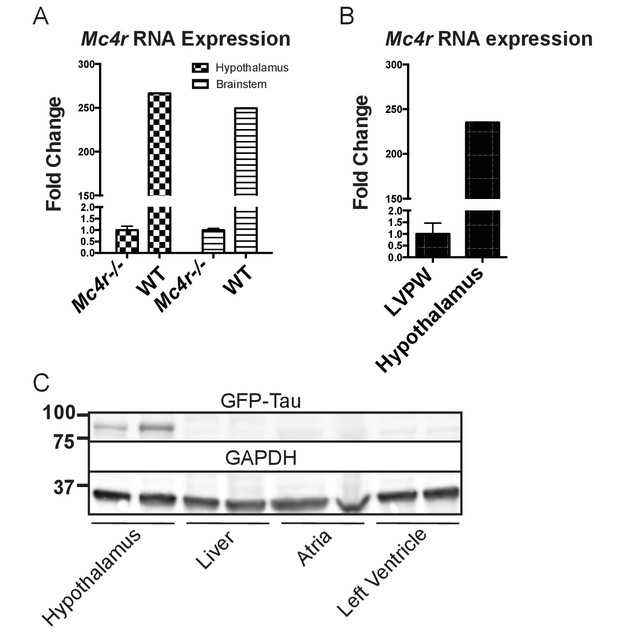
Mc4r is not expressed in adult heart tissue.
(A) qRT-PCR comparison of Mc4r expression in heart and hypothalamus using the ΔΔCT method. (Student’s t-test, p<0.0001, n = 8) (B) qRT-PCR comparison of Mc4r expression in the hypothalamus and brainstem between Mc4r−/− and WT tissue demonstrating dynamic range of qRT-PCR primers (Student’s t-test, p<0.0001, n = 8). (C) Representative western blot of Mc4r -Tau-Sapp hypothalamic, hepatic, atrial and ventricular lysates.
-
Figure 4—source data 1
Source data for Figure 4A–B.
This source file provides mean, SD, sample size and statistical test information for all qRT-PCR experiments in Figure 4.
- https://doi.org/10.7554/eLife.28118.009
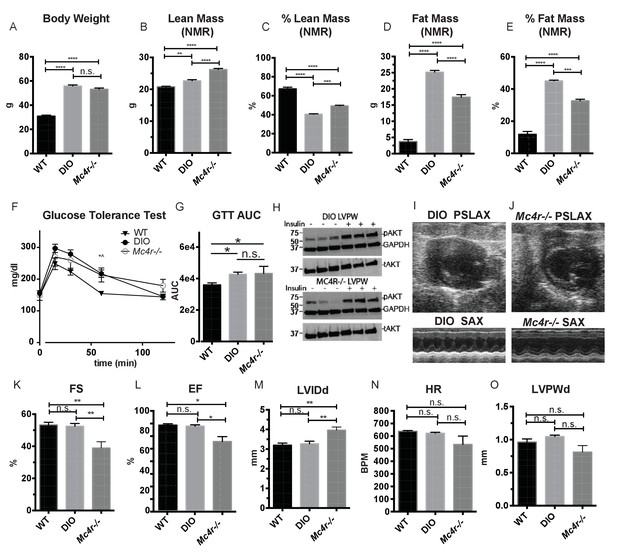
Age and weight matched wild type animals fed a high fat diet do not phenocopy the cardiomyopathy seen in Mc4r−/− mice.
(A) Body weights of age matched WT, diet induced obese (DIO), and Mc4r−/− mice. (1-way ANOVA Sidak multiple comparison test, p<0.0001, n = 5). (B) Lean mass of WT, DIO and Mc4r−/− mice by NMR (1-way ANOVA Sidak multiple comparison, p<0.01, n = 5) (C) Percent Lean Mass comparison of WT, DIO and Mc4r−/− mice (1-way ANOVA Sidak multiple comparison, p<0.001, n = 5) (D) Fat mass by NMR (1-way ANOVA Sidak multiple comparison, p<0.0001, n = 5) (E) Percent Fat Mass (1-way ANOVA Sidak multiple comparison, p<0.0001, n = 5) (F) Glucose tolerance test time course. 2 mg glucose per kg lean mass (2-way ANOVA Sidak multiple comparison test, p<0.05, n = 5) (G) Area under the curve of glucose tolerance test (Dunn’s multiple comparison test, p<0.05, n = 5) (H) pAkt T-308 blots of the LVPW in Mc4r−/− and DIO mice following the injection of saline or 1 U/kg insulin. (I) Representative B-mode PSLAX image at diastole (top) and M-mode SAX image of male DIO mice at 30 weeks of age. (J) Representative B-mode PSLAX image at diastole (top) and M-mode SAX image of male Mc4r−/− mouse 30 weeks of age. (K) Fractional Shortening of WT, DIO and Mc4r−/− mice (1-way ANOVA Sidak multiple comparison, p<0.01, n = 5) (L) Ejection Fraction of WT, DIO and Mc4r−/− mice (1-way ANOVA Sidak multiple comparison, p<0.05, n = 5) (M) LVIDd (1-way ANOVA Sidak multiple comparison, p<0.01, n = 5) (N) HR (1-way ANOVA Sidak multiple comparison, p>0.05, n = 5) (O) LVPWd (1-way ANOVA Sidak multiple comparison, p>0.05, n = 5).
-
Figure 5—source data 1
Source data for Figure 5A–G and K-O.
This source file provides mean, SD, sample size and statistical test information for all body compostion, glucose tolerance, and echocardiography studies in Figure 5.
- https://doi.org/10.7554/eLife.28118.011
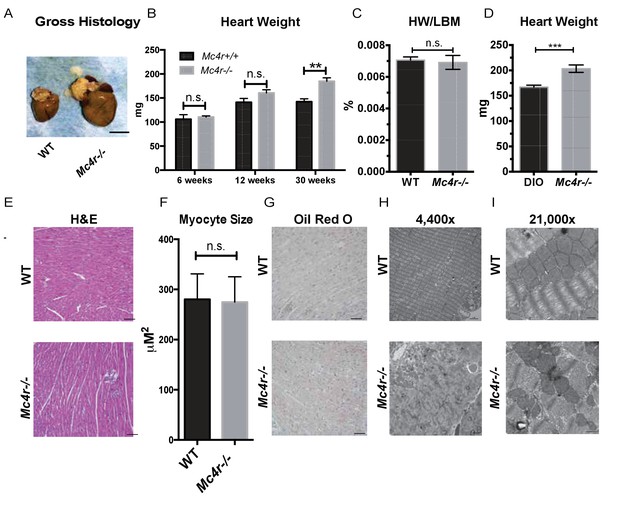
Histological analysis of Mc4r−/− hearts reveals mitochondrial pleomorphy and cardiomyocyte dropout.
(A) Representative gross pathology of WT and Mc4r−/− hearts (B) Heart wet weight of 6, 12 and 30 week old WT and Mc4r−/− animals (Student’s t-test, p<0.01, n = 4–7) (C) Heart weight to total lean mass ratio (Student’s t-test, p>0.05, n = 5) (D) Heart wet weight of 40 week old Mc4r−/− and DIO animals. (Student’s t-test; p<0.001, n = 7–8). (E) Representative H and E images of WT and Mc4r−/− myocardium; scale bar = 500 μm (F) Cross sectional myocyte area by H and E stain. (Student’s t-test, p>0.05, n = 3 mice x three cross sections). (G) Representative Oil Red O images of WT and Mc4r−/− myocardium; scale bar = 500 μm (H-I) Representative TEM images WT and Mc4r−/− myocardium; 4400x scale bar = 2 μm; 21,000x scale bar = 500 nm.
-
Figure 6—source data 1
Source data for Figure 6B–D and 6F.
This source file provides mean, SD, sample size and statistical test information for the gross pathological and histological analyses presented in Figure 6.
- https://doi.org/10.7554/eLife.28118.013
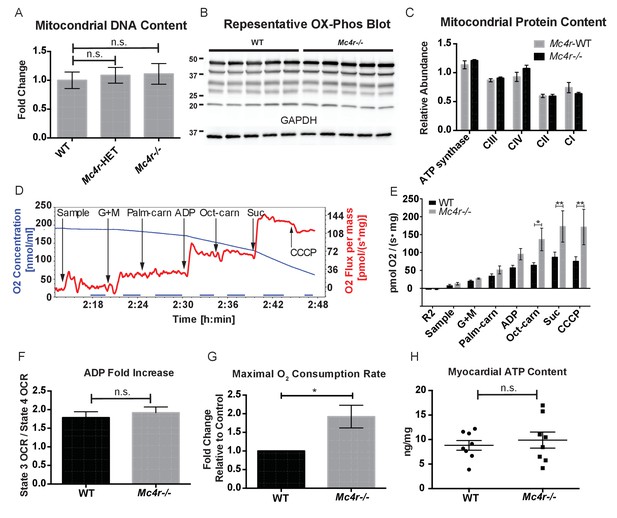
Mc4r−/− heart tissue displays increased mitochondrial capacity.
(A) Relative qPCR of mitochondrial DNA content using H19 and CytB primers (Students t-test of ΔCT values, p>0.05, n = 6). (B) Representative western blot of whole cell tissue lysates from WT and Mc4r−/− myocardium of electron transport chain (ETC) proteins. (C) Relative quantification of ETC blot normalized to GAPDH content (2 way ANOVA Sidak multiple comparison test, p>0.05, n = 3 blots x 5 samples per genotype) (D) Representative trace of a respirometry assay. Chamber O2 content is represented in blue while the rate of O2 consumption per mg tissue is in red. The sampling time used for quantification is displayed in the blue bar on the x-axis and is obtained following stabilization of O2 consumption rate in the chamber. (E) O2 consumption rate comparison between permeabilized WT and Mc4r−/− myocardial tissue from 30 to 33 week old mice. (2 way ANOVA Sidak post test, p<0.05, n = 5) (F) ADP dependent O2 consumption ratio (paired t-test, p>0.05, n = 5) (G) Ratio O2 consumption of Mc4r−/− tissue compared to WT (ratio paired t-test, p<0.05, n = 5) (H) ATP content in WT and Mc4r−/− myocardial tissue (Student’s t-test, p>0.05, n = 9).
-
Figure 7—source data 1
Source data for Figure 7A and C, and 7E-H.
This source file provides mean, SD, sample size and statistical test information for all studies presented in Figure 7.
- https://doi.org/10.7554/eLife.28118.015
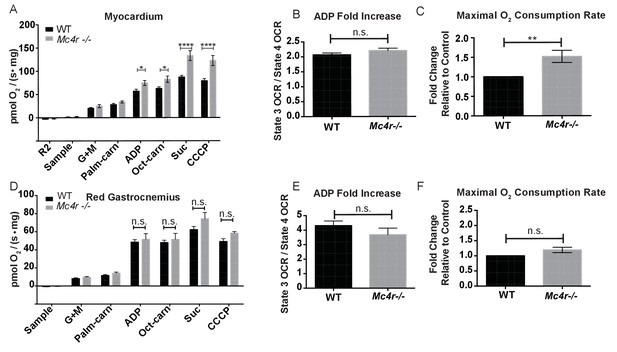
Mitochondrial capacity changes in Mc4r−/− animals are present in young animals and specific to the heart.
(A) O2 consumption rate comparison between WT and Mc4r−/− myocardial tissue from 6 to 12 week old mice. (2 way ANOVA Sidak post test, p<0.05, n = 7) at 6–12 weeks of age (B) ADP dependent O2 consumption ratio (paired t-test, p>0.05, n = 7) (C) Ratio O2 consumption of Mc4r−/− tissue compared to WT (ratio paired t-test, p<0.05, n = 7) (D) O2 consumption rate comparison between WT and Mc4r−/− red gastrocnemius tissue from 6 to 12 week old mice. (2 way ANOVA Sidak post test, p>0.05, n = 6) at 6–12 weeks of age (E) ADP dependent O2 consumption ratio of red gastrocnemius tissue from 6 to 12 week old mice (paired t-test, p>0.05, n = 6) (F) Ratio O2 consumption of Mc4r−/− red gastrocnemius tissue compared to WT red gastrocnemius tissue (ratio paired t-test, p>0.05, n = 6).
-
Figure 8—source data 1
Source data for Figure 8A–F.
This source file provides mean, SD, sample size and statistical test information for all high resolution respirometry studies in Figure 8.
- https://doi.org/10.7554/eLife.28118.017
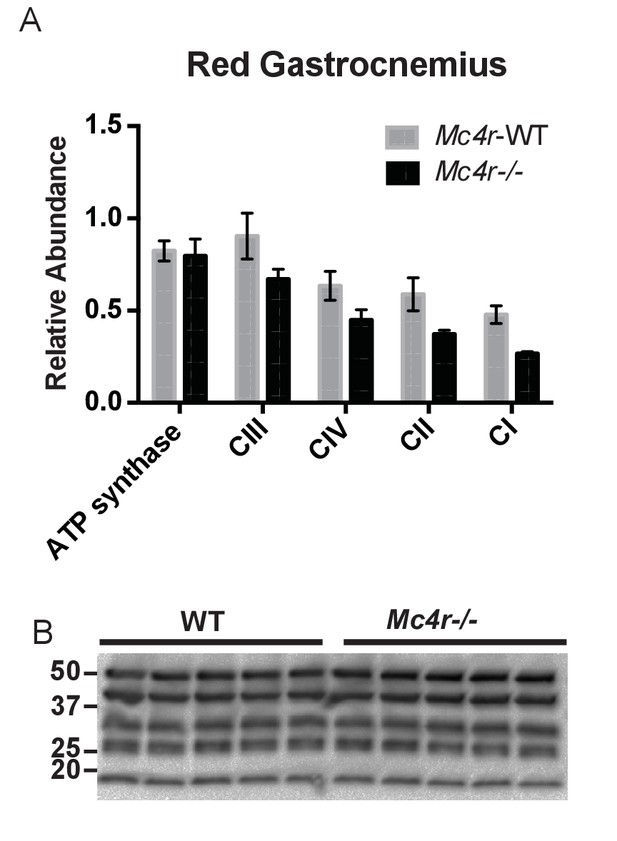
Red Gastrocnemius Ox-Phos components.
(A) Relative quantification of ETC blot normalized to ATP synthase content in WT tissue (2 way ANOVA Bonferroni post test, p>0.05, n = 3 blots x 5 samples per genotype) (B) Representative western blot from tissue lysates from WT and Mc4r−/− myocardium of ETC proteins.
-
Figure 9—source data 1
Source data for Figure 9A.
- https://doi.org/10.7554/eLife.28118.019
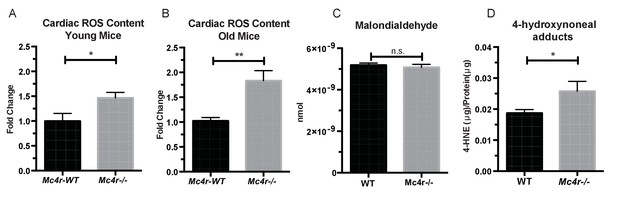
Mc4r−/− hearts display signs of increased ROS production.
(A) DCF assay comparing myocardial lysates from 6 to 12 week old Mc4r−/− and WT mice (ratio paired t-test, p<0.05, n = 5). (B) DCF assay comparing myocardial lysates from 30 to 33 week old Mc4r−/− and WT mice (ratio paired t-test, p<0.05, n = 5). (C) Malondialdehyde content in 30 week old Mc4r−/− and WT myocardium (Student’s t-test, p>0.05, n = 5) (D) 4-HNE protein content per mg total protein (Student’s t-test, p>0.05,n = 5).
-
Figure 10—source data 1
Source data for Figure 10A–D.
This source file provides mean, SD, sample size and statistical test information for all ROS studies presented in Figure 10.
- https://doi.org/10.7554/eLife.28118.021
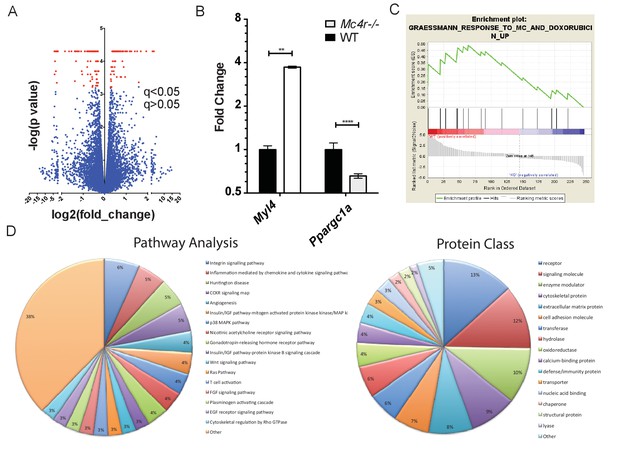
Gene changes in Mc4r−/− myocardium.
(A) Volcano plot of RNA-seq results. Red points indicate gene changes that reached statistical significance (n = 4 WT, n = 6 Mc4r−/−). (B) qRT-PCR confirmation of select gene changes found with RNA-seq (Student’s t-test, p<0.01, n = 4 WT, n = 6 Mc4r−/−) (C) Gene set enrichment analysis (GSEA) results from a different gene transcripts found using RNA-seq (nominal p<0.001). (D) Gene ontology pathway analysis of significantly unregulated genes using PANTHER Pathways Overrepresentation test.
-
Figure 11—source data 1
Source data for Figure 11B.
This source file provides mean, SD, sample size and statistical test information for the qRT-PCR studies presented in Figure 11B.
- https://doi.org/10.7554/eLife.28118.023
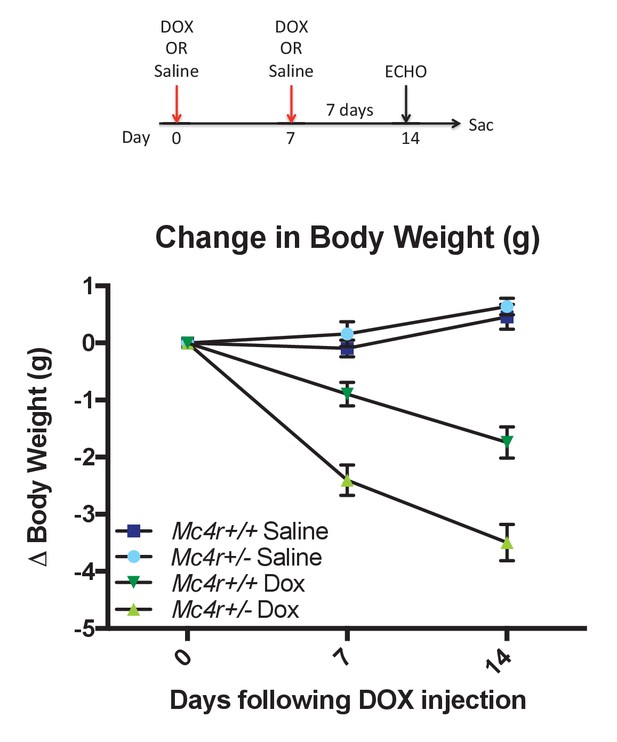
Mc4r ± mice are hypersensitive to doxorubicin.
(A) Treatment strategy to subclinical cardiac dysfunction in WT C57B6/J mice. Mice were given 5 mg/kg doxorubicin over the course of 2 weeks during which time their bodyweights were recorded. (B) Body weights of Mc4r+/- and WT during the course of the treatment paradigm. Mc4r+/- mice almost twice as much weight as their WT controls. (One Way ANOVA, p<0.05, n = 6–14).
-
Figure 12—source data 1
Source data for Figure 12B.
This source file provides mean, SD, sample size and statistical test information for the doxorubicin studies outlined in Figure 12A and presented in Figure 12B.
- https://doi.org/10.7554/eLife.28118.025
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28118.026





