Evidence that Mediator is essential for Pol II transcription, but is not a required component of the preinitiation complex in vivo
Figures
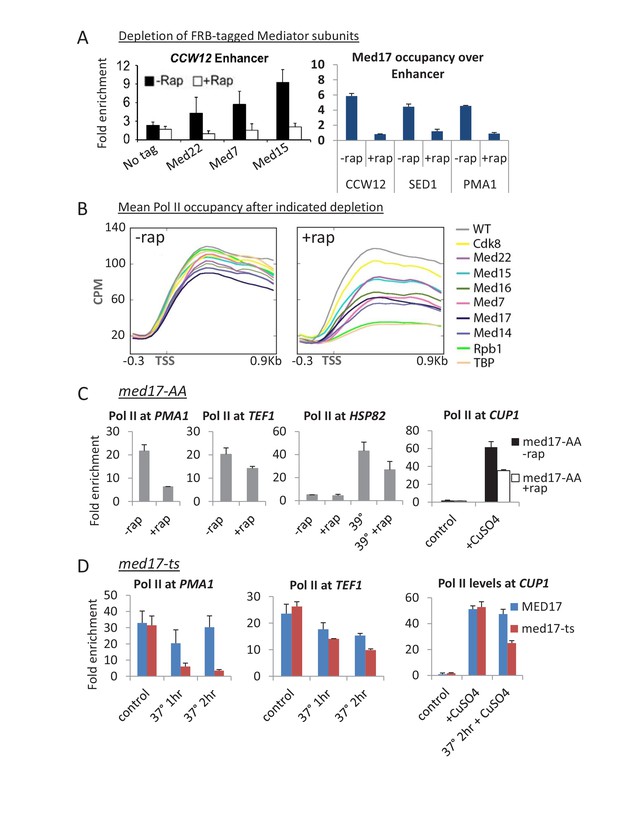
Substantial transcription persists upon anchor-away of essential Mediator subunits.
(A) Occupancy of the indicated 3x-HA-FRB tagged Mediator subunit at the indicated enhancers prior to (-Rap) or after (+Rap) being depleted by anchor away. The parental strain containing no FRB-tagged protein was used as a negative control. The HA antibody was used except for Med17, in which case an antibody to the native protein was used. Data from (Petrenko et al., 2016). (B) Mean Pol II occupancy over ~400 transcribed genes prior to and after anchor-away of the indicated Mediator subunits, TBP, Pol II (Rpb1) and the parental strain (WT). Sequence reads were normalized as counts per million (CPM), and the curves were aligned relative to the transcription start site (TSS). (C) Pol II occupancy at the indicated constitutive and induced (by heat shock at 39°C or addition of copper) genes prior to and after Med17 anchor-away. (D) Pol II occupancy at constitutive genes and at the copper-inducible CUP1 gene prior to and after heat inactivation of a med17-ts allele. An isogenic MED17 strain was used as the control.
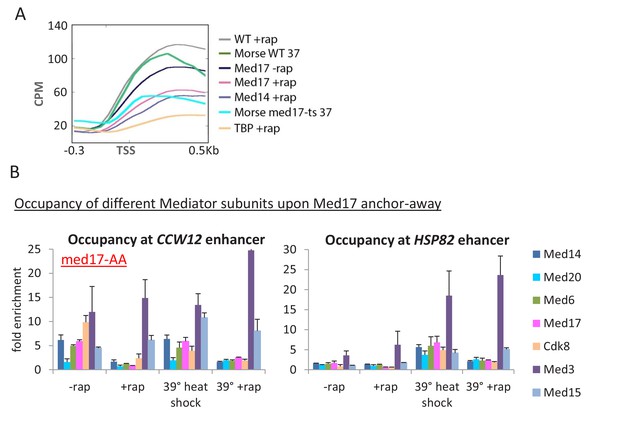
Depletion of Med17 function via the ts mutant or anchor away results in similar transcriptional effects.
(A) Mean Pol II occupancy over ~ 400 transcribed genes prior to and after anchor-away of Med14, Med17, TBP, or the parental strain (WT); data is the same as shown in Figure 1B. Pol II data for the med17-ts and parental strain at 37°C from (Paul et al., 2015) was downloaded and analyzed in the same manner. Sequence reads were normalized as counts per million (CPM), and the curves were aligned relative to the transcription start site (TSS). (B) Occupancy of the indicated Mediator subunits and Pol II at the CCW12 and HSP82 enhancers of the indicated genes in a med17-AA strain that was or was not treated for 1 hr with rapamycin and was or was not subjected to a 15 min heat shock at 39°C. Data from (Petrenko et al., 2016).
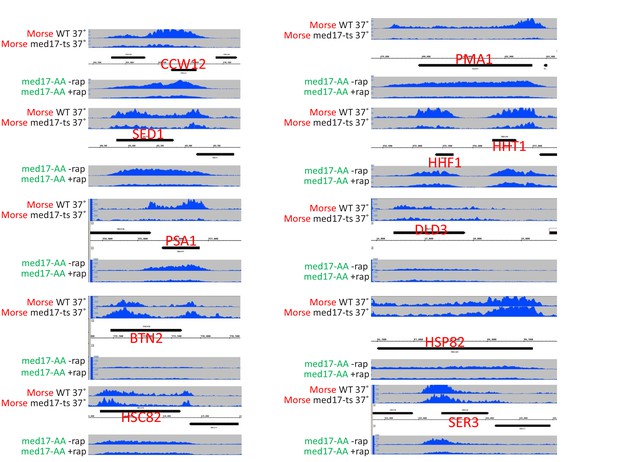
Screenshots of Pol II occupancy for the indicated genes for samples analyzed in Figure 1—figure supplement 1A.
https://doi.org/10.7554/eLife.28447.004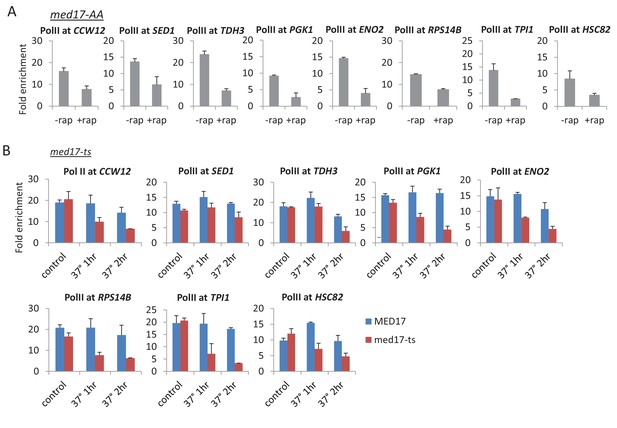
Pol II occupancy at the indicated constitutive genes in the (A) Med17-AA (± rapamycin) and (B) med17-ts (at permissive temperature and 37°C for 1 or 2 hr) strains.
https://doi.org/10.7554/eLife.28447.005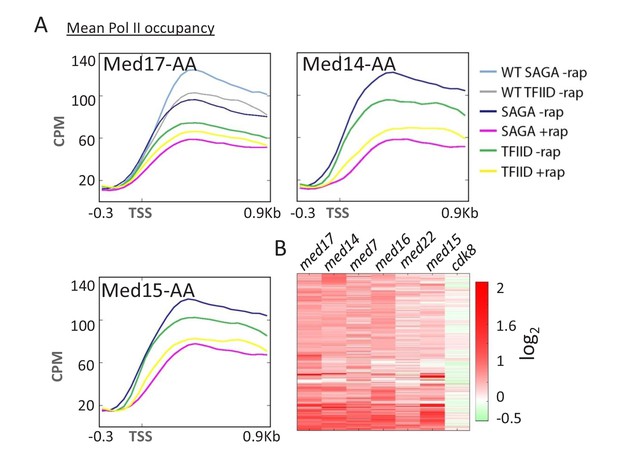
Mediator preferentially affects SAGA-dependent vs.
TFIID-dependent genes. (A) Mean Pol II occupancy for the indicated groups of genes (SAGA or TFIID-dependent) prior to and after anchor-away of Med17, Med14, and Med15, as well as for the parent strain (WT). Sequence reads were normalized as counts per million (CPM), and the curves were aligned relative to the transcription start site (TSS). (B) Heat map of Pol II levels (log2 scale) at individual genes before vs. after anchor-away of the indicated Mediator subunits. Occupancy changes upon anchor-away are indicated as decreases (red) or increases (green).
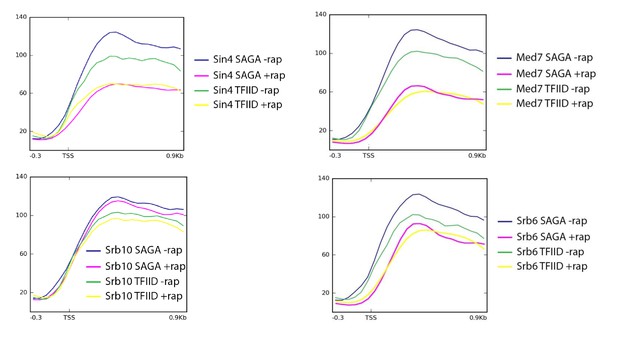
Mean Pol II occupancy for the indicated groups of genes (SAGA or TFIID-dependent) prior to and after anchor-away of Sin4 (Med16), Srb6 (Med22), Srb10 (Cdk8), and Med7.
Sequence reads were normalized as counts per million (CPM), and the curves were aligned relative to the transcription start site (TSS).
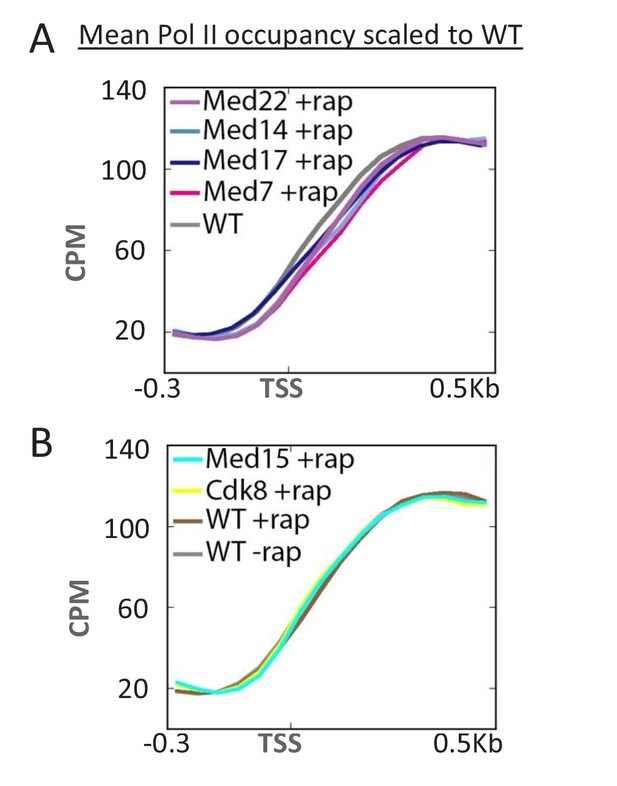
Depletion of Mediator head and middle subunits causes a downstream shift in the Pol II profile.
(A) Overlaid mean Pol II occupancy curves scaled to 100% (maximum levels) after anchor-away of the indicated head and middle subunits of Mediator. (B) Overlaid mean Pol II occupancy curves scaled to 100% after anchor-away of the indicated tail and kinase module subunits, as well as for the parent strain (WT) before and after rapamycin addition. Sequence reads were normalized as counts per million (CPM), and the curves were aligned relative to the transcription start site (TSS).
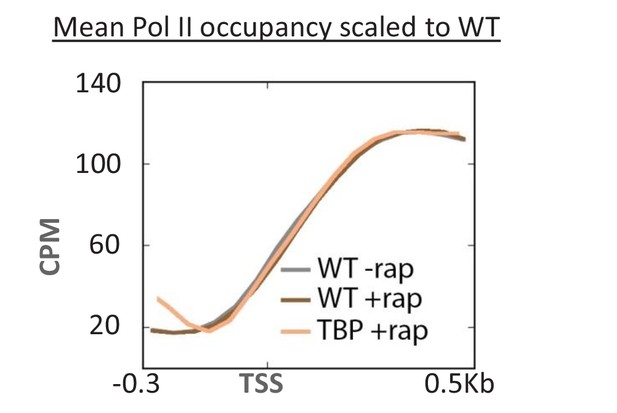
Overlaid mean Pol II occupancy curves scaled to 100% after anchor-away of TBP, as well as for the parent strain (WT) before and after rapamycin addition.
Sequence reads were normalized as counts per million (CPM), and the curves were aligned relative to the transcription start site (TSS).
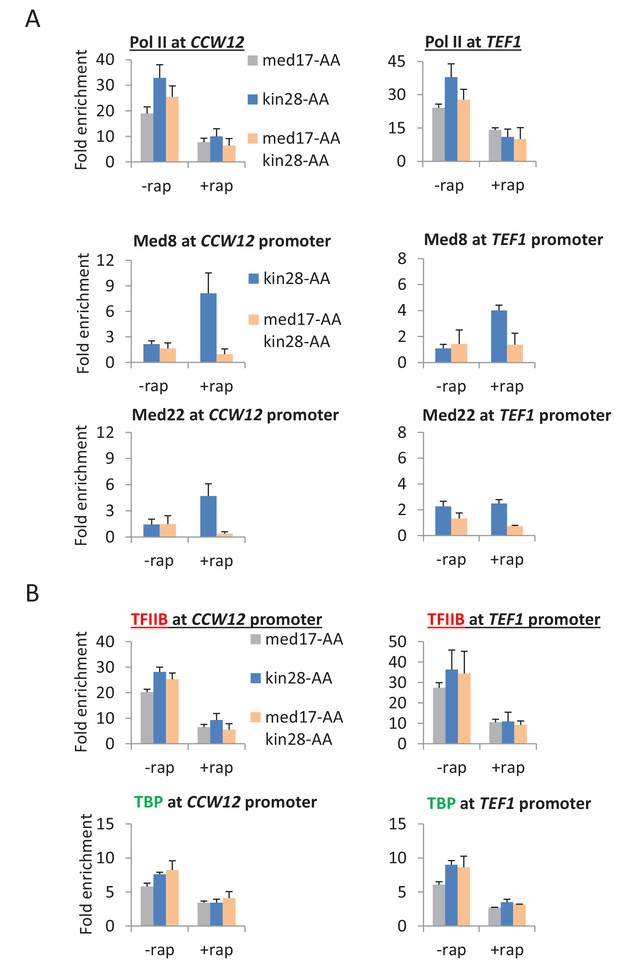
Pol II transcription can occur from preinitiation complexes lacking Mediator.
(A) Pol II occupancy in the coding regions and Mediator subunits at the promoters of the CCW12 and TEF1 genes, before or after anchor-away of Med17, Kin28, or both simultaneously. (B) TBP and TFIIB occupancy at the CCW12 and TEF1 promoters in the same samples.
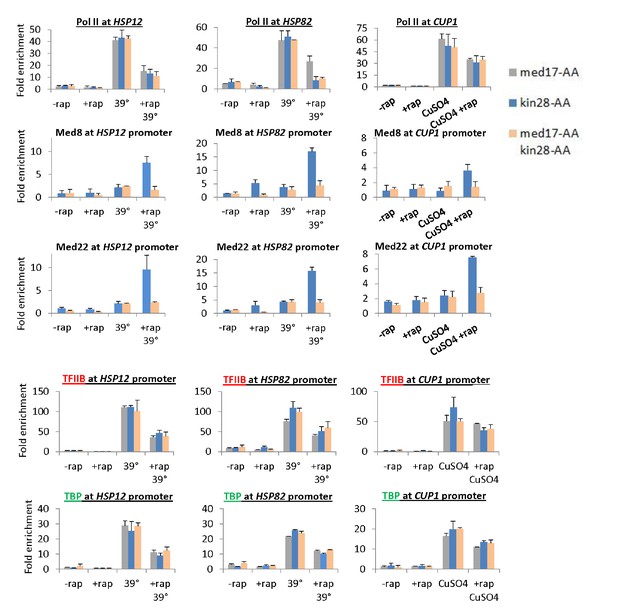
Pol II transcription can occur from preinitiation complexes lacking Mediator.
(A) Occupancy of Pol II in the coding regions, and Mediator (Med8 and Med22 subunits), TBP, and TFIIB subunits at the promoters of the inducible HSP12, HSP82, and CUP1 genes before or after anchor-away of Med17, Kin28, or both simultaneously. HSP12 and HSP82 were analyzed before and after a heat shock at 39°C, whereas CUP1 was analyzed before and after the addition of copper.
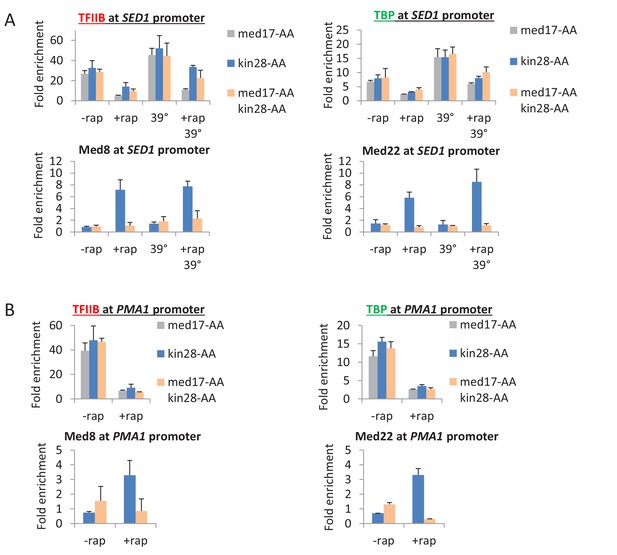
Pol II transcription can occur from preinitiation complexes lacking Mediator.
Occupancy of Mediator (Med8 and Med22 subunits), TBP, and TFIIB at the promoters (A) SED1 and (B) PMA1 in the presence of absence of rapamycin and the presence or absence of a heat shock at 39°C.
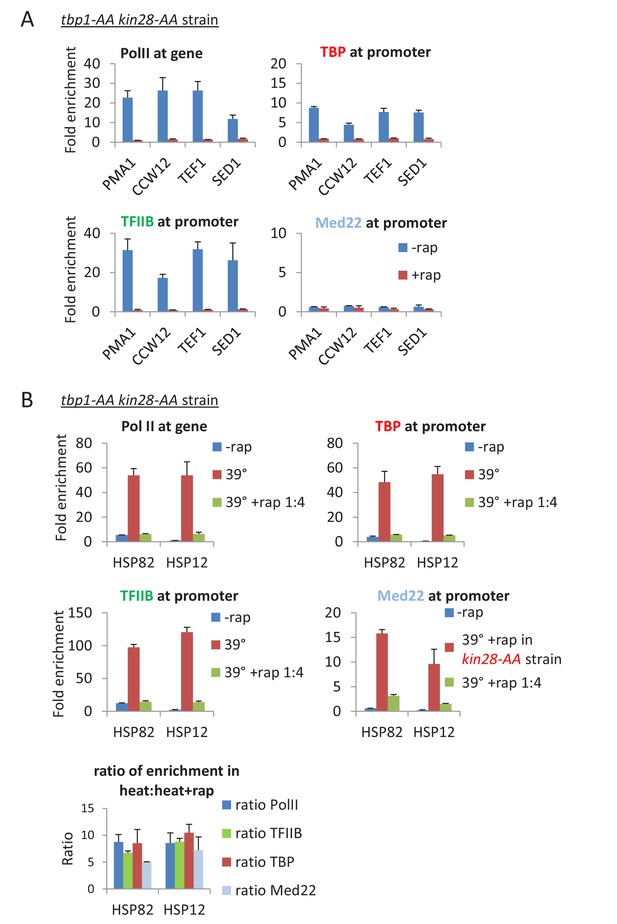
Depletion of TBP does not alter the composition of the preinitiation complex.
(A) Occupancy of Pol II in the coding regions, and Med22, TBP, and TFIIB at the promoters of the indicated genes before or after simultaneous depletion of TBP and Kin28. (B) Occupancy of Pol II in the coding regions, and Med22, TBP, and TFIIB at the promoters of the indicated heat shock genes before or after a heat shock at 39°C in cells that were or were not incompletely depleted for TBP and Kin28 by using rapamycin at 25% of the usual concentration. As Mediator can only be detected at the core promoter upon Kin28 depletion, Med22 occupancy at heat shock promoters were tested in a kin28-AA strain. The relative occupancy ratios of Pol II, TBP, TFIIB, and Med22 in heat shocked cells that were or were not incompletely depleted is shown below.
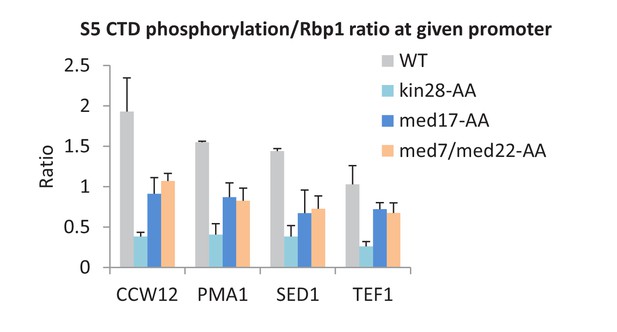
Mediator is important, but not essential, for serine 5 phosphorylation of the Pol II CTD.
The ratio of S5-phosphorylated CTD levels relative to total Rpb1 levels at the promoters of the indicated genes, depletion of the indicated proteins or the control strain (WT).
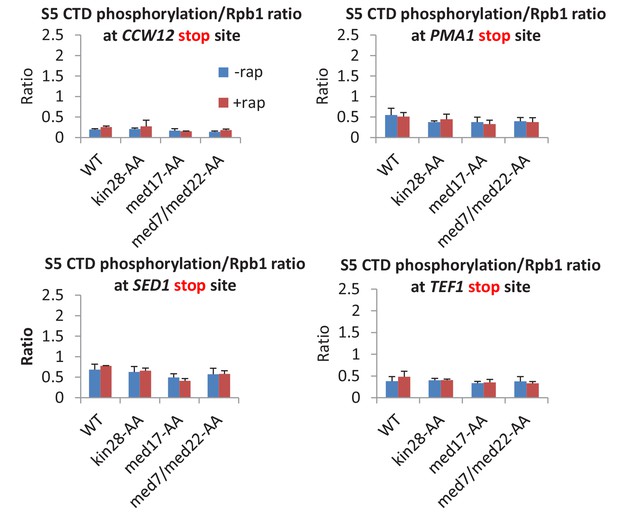
The ratio of S5-phosphorylated CTD levels relative to Rpb1 levels at the transcription stop sites of the indicated genes prior to or after Kin28 anchor-away, Med17 anchor-away, or a double anchor-away of Med7 and Med22, as well as in the parent strain (WT).
https://doi.org/10.7554/eLife.28447.015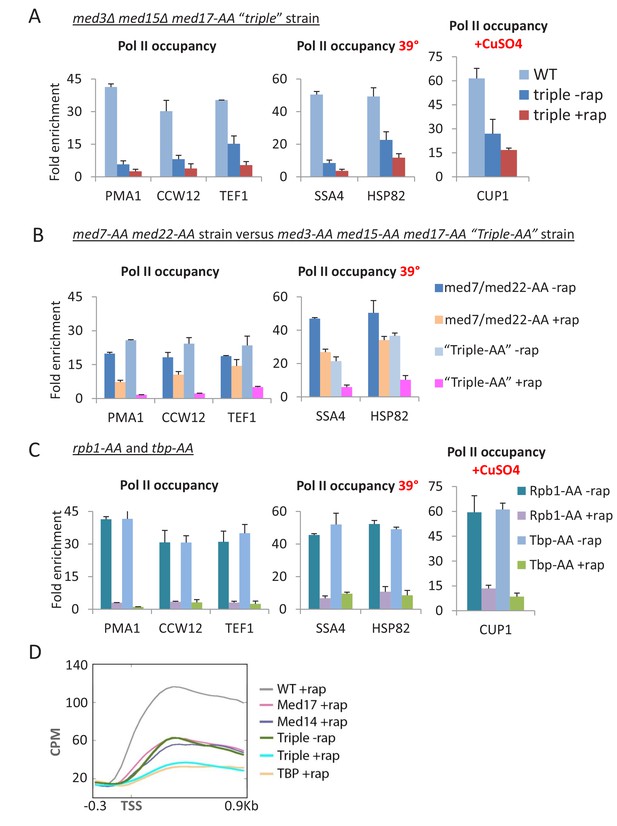
Pol II transcription is virtually eliminated when Mediator head, middle, and tail modules are simultaneously inactivated.
(A) Pol II occupancy in the coding regions of the indicated constitutive and inducible genes for the parent strain (WT) and ‘triple’ mutant strain, in which the tail module is inactivated via deletion of med3 and med15, and the head and middle modules are inactivated via anchor-away of Med17. (B) Pol II occupancy in the coding regions of the indicated constitutive and inducible genes for the parent strain (WT) and a strain simultaneously depleted for Med3, Med15 (tail module), and Med17 (head module) or for Med7 (middle module) and Med22 (head module). (C) Pol II occupancy in the same genes prior to and after anchor-away of Rpb1 and TBP. (D) Mean Pol II occupancy over ~400 transcribed genes in strains depleted for Med14 or Med17 as well as the triple mutant strain (before and after rapamycin) and the parental strain (WT). Sequence reads were normalized as counts per million (CPM), and the curves were aligned relative to the transcription start site (TSS).
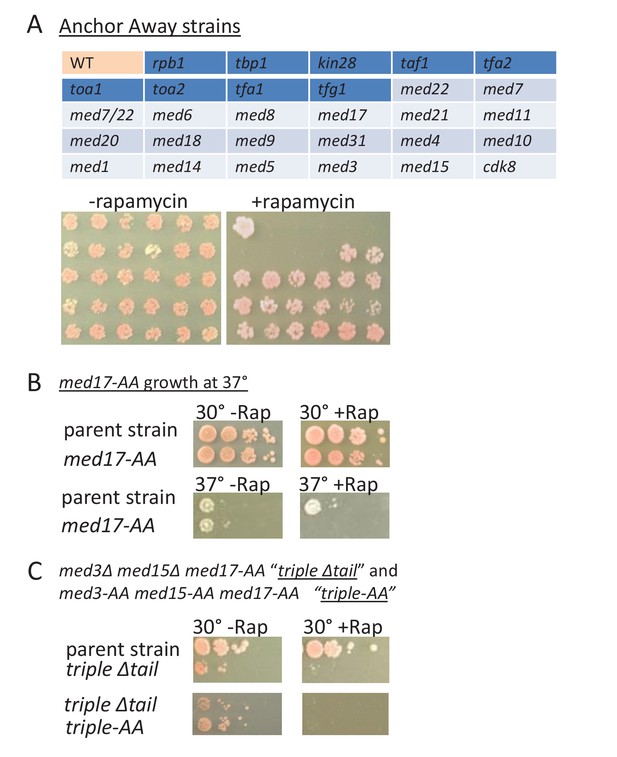
Growth of Mediator depletion strains.
(A) growth on YPD (2.5 days) in the presence or absence rapamycin upon anchor-away mediated depletion of the Mediator subunit (light blue shading) or general transcription factor (dark blue shading) corresponding to the key in the upper panel. WT corresponds to the parent strain. (B) Growth of the med17-anchor-away strain and of the parent strain on YPD at 30° or 37° in the presence or absence rapamycin. (C) Growth of the parental, ‘triple’ mutant, and triple depletion strain, on YPD at 30° in the presence or absence of rapamycin.
Additional files
-
Supplementary file 1
List of strains.
- https://doi.org/10.7554/eLife.28447.018





