Somatic hypermutation of T cell receptor α chain contributes to selection in nurse shark thymus
Figures

Alignment of Gamma V clones suggests minimal somatic hypermutation.
Thymocyte clones for nine γV groups from three different predicted V genes. CDR regions are marked above the scale for each γV alignment. Amino acids are shown under the nucleotide consensus sequence, and dots represent identity to this sequence. We highlighted nonsynonymous changes in black; synonymous changes are underlined. Gaps are used for alignment purposes and indicate a shortened sequence (at the beginning or end). Sequences are identified by a single clone number or a group of identical clones condensed to a single line (the number of clones are indicated). Clone numbers that contain ‘THY’ are from thymus, ‘PBL’ are from peripheral blood leukocytes, and ‘SPV’ are from spiral valve (intestine). We deposited all 69 sequences into GenBank under accession numbers KY351639 – KY351707.

Alignment of Delta V clones suggests somatic hypermutation.
Thymocyte clones for 12 δV groups from seven different predicted V genes. CDR regions are marked above the scale for each δV alignment. Amino acids are shown under the nucleotide consensus sequence, and dots represent identity to this sequence. We highlighted nonsynonymous changes in black; synonymous changes are underlined. Gaps are used for alignment purposes and indicate a shortened sequence (at the beginning or end). Sequences are identified by a single clone number or a group of identical clones condensed to a single line (the number of clones are indicated). Clone numbers that contain ‘THY’ are from thymus, ‘PBL’ are from peripheral blood leukocytes, and ‘SPV’ are from spiral valve (intestine). We deposited all 112 sequences into GenBank under accession numbers KY346705 – KY346816.

Alignment of Beta V clones illustrates a lack of somatic hypermutation.
Thymocyte clones for βV groups from six different predicted V genes. CDR regions are marked above the scale for each βV alignment. Amino acids are shown under the nucleotide consensus sequence, and dots represent identity to this sequence. We observed three nonsynonymous changes within a single sequence (highlighted in black). Sequences are identified by a single clone number or a group of identical clones condensed to a single line (the number of clones are indicated). Clone numbers that contain ‘PBL’ are from peripheral blood leukocytes and ‘THY’ are from thymus. We deposited all 57 sequences into GenBank under accession numbers KY351708 – KY351764.

CDR3s of TcR Alpha chain are diverse.
Amino acid (aa) alignment of TcR αV1 thymocyte clones illustrating diversity of the third complementarity-determining region (CDR3). All clones contain identical variable (V) region sequence (aa 1–61). We grouped clones by shared, identical joining (J) regions (purple boxes) and highlight the differences in the V-J join (CDR3 region) in red boxes.
-
Figure 4—source data 1
CDR3 regions diversified by exonuclease activity and addition of N and P nucleotides.
Alignment of nucleotides belonging to the join between variable (V) and joining (J) segments within TcRα thymocyte clones. We determined the putative ends of each V segment and putative beginning of each J segment by comparing alignments between different sharks, assuming that identical nucleotides between sharks were germline. The last number of each sequence name indicates the number of clones containing that nucleotide sequence between the V and J segments.
- https://doi.org/10.7554/eLife.28477.006
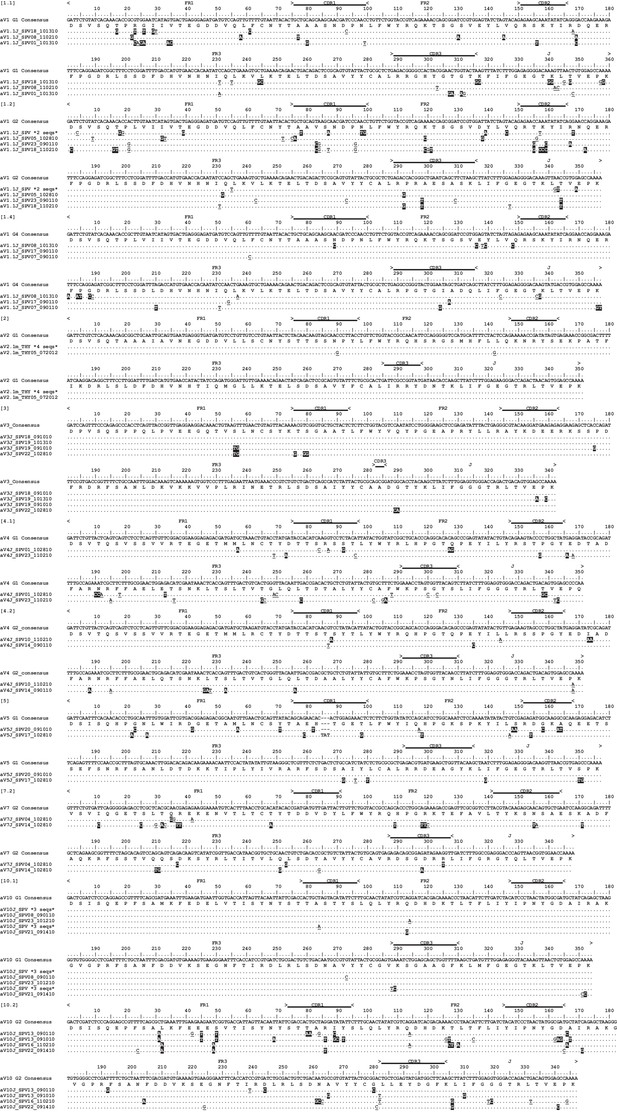
Alignment of Alpha V cDNA clones suggest somatic hypermutation at shark TCRα.
Thymocyte clones for all 11 αV groups with the same CDR3 from six different predicted V genes. Locations of framework regions (FR), complementarity determining regions (CDR), joining regions (J), and constant (C) regions are marked above the scale for each αV. In absence of germline sequence information, we used a Geneious-derived nucleotide consensus sequence for analysis of nucleotide changes in thymocyte clones. Amino acids are shown under the consensus sequence, and dots represent identity to this sequence. Nonsynonymous changes are highlighted in black; synonymous changes are underlined. Gaps are used for alignment purposes and indicate either a shortened sequence (at the beginning or end) or insertions or deletions within the sequence. Sequences are identified by a single clone number or a group of identical clones condensed to a single line (the number of clones are indicated). Clone numbers that contain ‘THY’ are from thymus, ‘PBL’ are from peripheral blood leukocytes, and ‘SPV’ are from spiral valve (intestine). We did not use clones from αV1.3 and αV7.1 because they did not contain mutations in FR or CDR regions. We deposited all 42 sequences into GenBank under accession numbers KY189332 – KY189354 or KY366469 – KY366487.
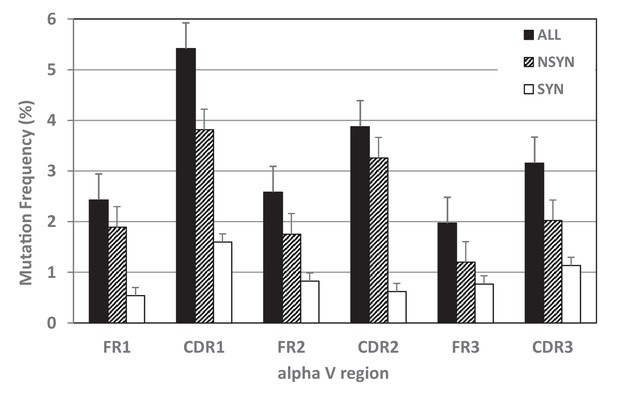
Mutation frequencies differed within TcR V regions.
Mutability of complementarity determining regions (CDR), especially CDR1, exceeded that of framework regions (FR) for all mutations together (black bars) and for nonsynonymous mutations alone (NSYN, hatched bars). We found no statistical difference in synonymous mutations (SYN, white bars) between CDRs and FRs.
-
Figure 6—source data 1
Frequency of mutation in TcR alpha V framework and complementarity determining regions for all mutation, nonsynonymous (NSYN) mutation only, or synonymous (SYN) mutation only.
- https://doi.org/10.7554/eLife.28477.013
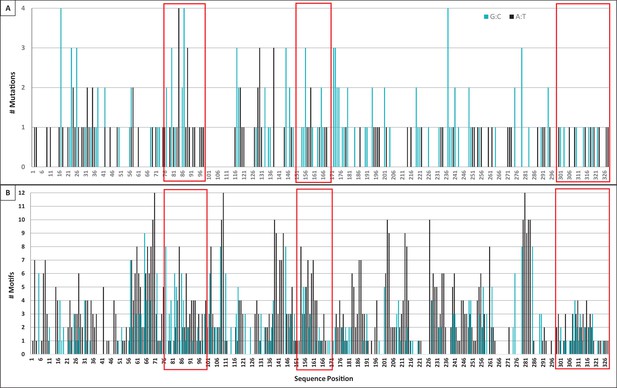
Mutation and motif locations within individual domains of TcR αV sequences.
(A) Number of mutations (both single and tandem, 283 total) to A:T nucleotides (black bars, 122 mutations) or G:C nucleotides (blue bars, 161 mutations) observed at each position along the sequence length of TcR αV sequences. We counted mutations to Geneious-derived consensus sequences within framework and complementarity-determining region (CDR) domains (41 sequences within 11 αV groups). (B) Number of WA/TW motifs (black bars, indicating possible polymerase η action) or DGYW/WRCH motifs (blue bars, indicating hotspots for AID activity, and thus possible mutation) at each position along the sequence length. Position indicates the forward (3’ to 5’) location of the mutable base of each motif within a Geneious-derived consensus sequence for each TcR αV group (eleven groups). Red boxes indicate the location of CDR1 (positions 76–99), CDR2 (positions 151–171), and CDR3 (positions 300–328) within each panel. [WA/TW: A:T is the mutable position; DGYW/WRCH: G:C is the mutable position; W = A/T, D = A/G/T, Y = C/T, R = A/G, and H = T/C/A].
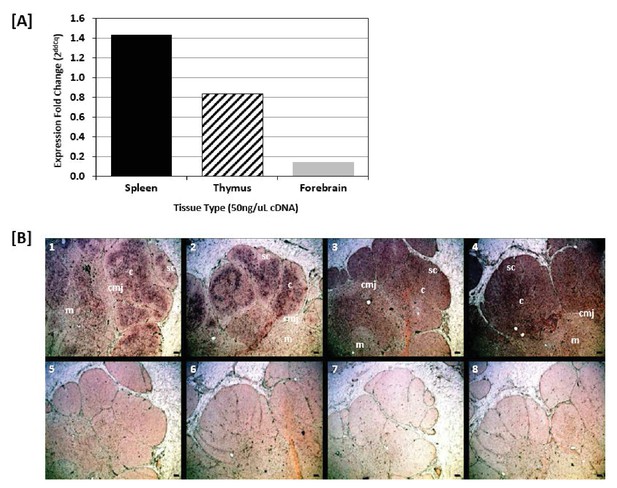
Shark thymus expresses AID.
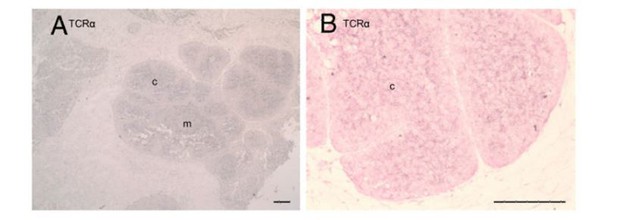
(A) Expression of AID in shark spleen (black bar), thymus (hashed bar), and forebrain (gray bar) using real-time quantitative PCR. Expression was measured using the delta delta Cq method and are normalized against shark muscle tissue using β2M as a reference gene. Data represent expression fold-change differences at four cDNA concentrations. (B) In situ hybridization of mRNA using ribo-probes on adult shark thymus sections. 1,2: Two different fields probing for TcRβ antisense. 3,4: AID antisense. 5,6: TcRβ sense. 7,8: AID sense. All micrographs at 10X magnification; black scale bar in lower right of each panel is 100 uM. Anatomical structures are designated on the top panels [c: cortex; m: medulla; sc: subcapsular region; cmj: corticomedullary junction].
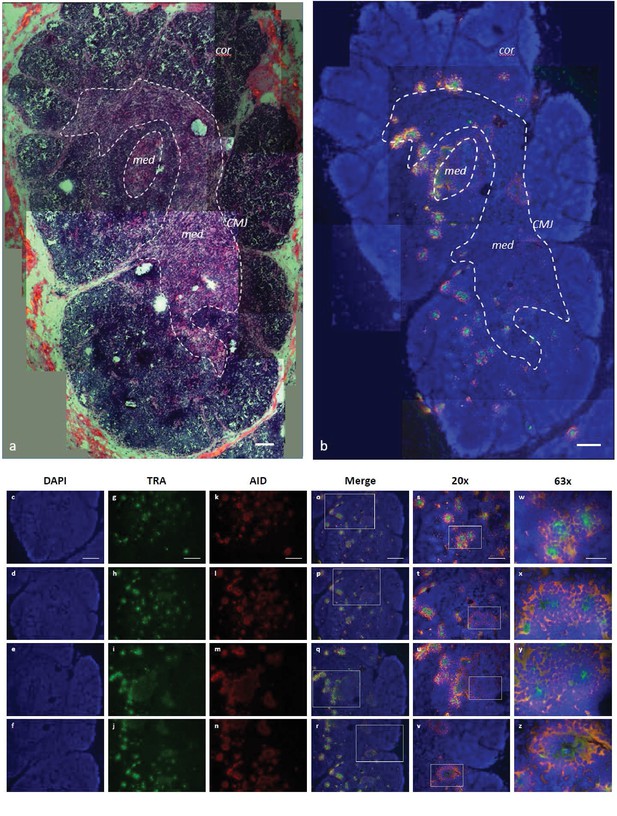
AID expression localized to inner cortex and cortico-medullary junction.
(a) H and E staining of fixed shark thymus tissue illustrating thymic architecture (10x). The densely packed cells at the margins of the image comprise the cortex (cor), while the less densely packed cells in the center constitute the medulla (med). The region at the junction between cortex and medulla incorporates the corticomedullary junction (CMJ), delineated generally by a hashed white circle. [b-z] Single molecule RNA fluorescence in situ hybridization (FISH) probing fixed thymus sections simultaneously for AID (probes labeled with Quasar 670; pseudo colored red) and TcRα (probes labeled with CalFluor Red 610; pseudo colored green) and counterstained with DAPI (blue). (b) Composite of seven Z-stacked images (10x) depicting overall thymic architecture and the localization of AID expression to the inner cortex and cortico-medullary junction regions of shark thymus. We superimposed (and minimally adjusted) the outlined CMJ boundaries from (a) onto (b) to elucidate the junction between cortex and medulla. [c-z] We obtained images of each fluorophore using 10x, 20x, and 63x magnification and merged Z-stacked images together. Individual (10x) fluorophore images of DAPI [c-f], TcRα [g-j], and AID [k-n] and Z-stacked merged images [o-r] illustrate AID and TcRα expression in four locations of shark thymus. White boxes indicate the magnified regions of the 10x and 20x images shown in the 20x [s-v] and 63x [w-z] images, respectively. Scale bars [a,b,c,g,k,o] 150 μm, [s] 75 μm, and [w] 30 μm. [cor: cortex; med: medulla; CMJ: corticomedullary junction].
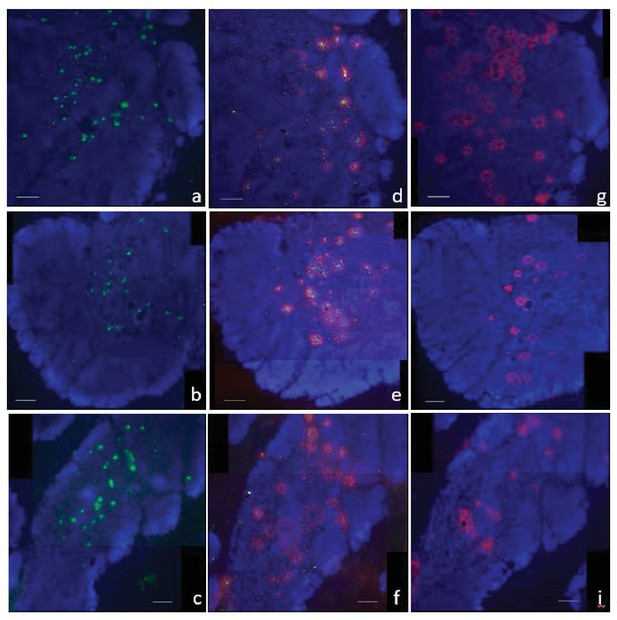
Localization of AID and TcRα probes is independent.
Single molecule RNA fluorescence in situ hybridization (FISH) showing three separate regions of fixed thymus sections demonstrating the independent localization of our probes. We probed individually for TcRα (a-c) (probes labeled with CalFluor Red 610; pseudo colored green), simultaneously for both TcRα and AID (d-f), or individually for AID (probes labeled with Quasar 670; pseudo colored red) and counterstained with DAPI (blue). There were two tissue sections in between imaged regions of TcRα section (a-c) and combined AID/TcRα section (d-f). The combined AID/TcRα section (d-f) was consecutive with the AID only section (g-h). We obtained images of each fluorophore using 10x magnification and merged Z-stacked images together. Scale bar 100 μm.
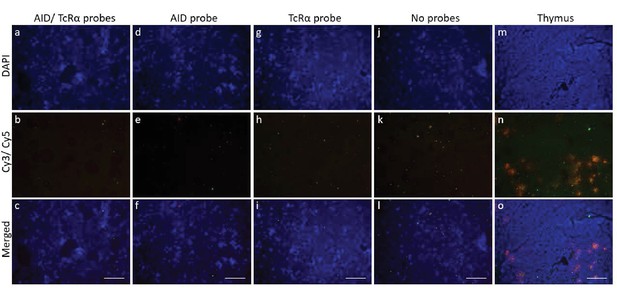
Lack of AID and TcRα probe hybridization in shark brain.
Single molecule RNA fluorescence in situ hybridization (FISH) showing fixed brain sections on four separate slides demonstrating the lack of probe localization in non-immune tissue. We probed simultaneously for both TcRα and AID (a-c), individually for AID (probes labeled with Quasar 670 and imaged with Cy5 filter; pseudo colored red) (d-f), individually for TcRα (probes labeled with CalFluor Red 610 and imaged with Cy3 filter; pseudo colored green) (g-i), or for neither probe (negative control) (j-l). We show probe staining in shark thymus tissue as a positive control [m-o]. We counterstained all slides with DAPI (blue). We imaged fluorescence detected using each filter (DAPI, Cy3/Cy5) regardless of the probe used to illustrate that background fluorescence is not dependent on probe application. We obtained images of each fluorophore using 10x magnification and merged Z-stacked images together (DAPI fluorescence [a,d,g,j,m]; Cy3 (green) and Cy5 (red) fluorescence [b,e,h,k,n]; merged [c,f,I,l,o]). Scale bar 150 μm.

Model predicting how AID acts on T cells in the thymus.
CD4/CD8 double negative (DN) thymocytes in the subcapsular region (SC) and cortex rearrange the β chain, using a surrogate pTα receptor to test for expression signaling. Cells with productive β arrangements then proliferate and express both CD4 and CD8, becoming double positive thymocytes (DPs). As DPs move toward the inner cortex and cortico-medullary junction (CMJ) where α chain rearranges, cells may begin to express AID. Non-productive rearrangements can be rescued from apoptosis by receptor editing or by receptor salvaging, in which AID catalyzes SHM to produce cells with improved affinity to MHC:Ag complexes (to pass positive selection). Salvaged thymocytes then proliferate and express either CD4 or CD8 on their surface as single-positive (SP) cells. AID-mediated receptor salvaging may also reduce recognition of self-peptide, rescuing self-reactive thymocytes from apoptosis (to pass negative selection). [sc: subcapsular region; cmj: corticomedullary junction; green shading indicates region of AID expression].
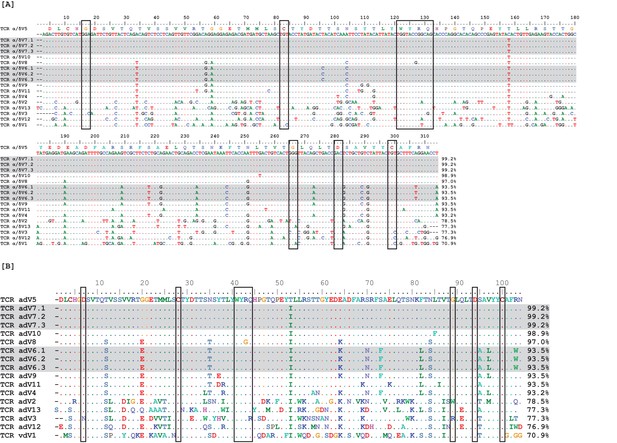
Observed TcR Alpha/Delta germline Vs exhibit high sequence identity.
Nucleotide (A) and amino acid (B) alignments of 17 germline variable (V) region gene segments. Two V groups contained three identical germline gene segments each (highlighted in gray), leaving only 13 unique V gene segments. Boxes surround conserved amino acids. Numbers at the ends of sequences indicate percent identity to the first germline sequence (α/δ V5) within the alignment.
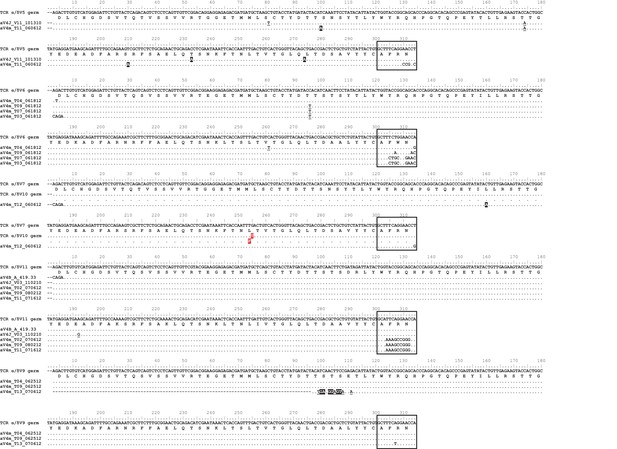
Observed germline sequences align only to TcR αV4 clones.
Nucleotide alignments of TcR αV4 thymocyte clones to known germline V segments. Highlighted and underlined bases denote nonsynonymous and synonymous differences (respectively) to the germline V segment. Boxed regions represent nucleotides of the third complementarity-determining region (CDR3) according to IMGT guidelines, accounting for differences between clone and germline sequences. We highlight the single nucleotide/amino acid change between α/δ V7 and α/δ V10 germline segments in red.
Tables
Summary of sequence data used in this paper.
Putative subfamilies within each TcR alpha V family share at least 85% nucleotide identity using nearest-neighbor consensus trees of V segments. Number of TcR alpha nucleotide (NUC), amino acid (AA) sequences or sequence groups within each category. Highlighted columns specifically refer to data used in this study. (See results for detailed descriptions of sequences included within each column.)
| Alpha V segment | Putative # Sub-families | All cloned sequences | Complete CDR3-J junction* | Unique V Region † | Unique V segment‡ | Unique CDR3-J§ | Groups with identical CDR3-J# | CDR3-J groups in Study** | Sequences in each dataset†† | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NUC | AA | NUC | AA | NUC | AA | |||||||
| TRA V1 | 1 | 40 | 40 | 35 | 34 | 24 | 16 | 34 | 32 | 7 | 4 | 2, 3, 3, 5 |
| TRA V2 | 3 | 18 | 18 | 13 | 13 | 13 | 10 | 12 | 12 | 3 | 1 | 5 |
| TRA V3 | 3 | 217 | 194 | 55 | 52 | 35 | 34 | 51 | 50 | 5 | 1 | 4 |
| TRA V4 | 3 | 60 | 28 | 22 | 22 | 21 | 21 | 21 | 21 | 6 | 2 | 2, 2 |
| TRA V5 | 4 | 35 | 34 | 15 | 13 | 9 | 8 | 13 | 13 | 3 | 1 | 2 |
| TRA V6 | 2 | 9 | 9 | 7 | 7 | 5 | 6 | 7 | 7 | 1 | 0 | 0 |
| TRA V7 | 5 | 96 | 60 | 49 | 48 | 39 | 38 | 48 | 48 | 9 | 2 | 2, 2 |
| TRA V9 | 3 | 19 | 19 | 14 | 14 | 12 | 11 | 13 | 13 | 4 | 0 | 0 |
| TRA V10 | 2 | 45 | 45 | 29 | 26 | 21 | 19 | 27 | 26 | 10 | 2 | 4, 9 |
| 539 | 447 | 239 | 229 | 179 | 163 | 226 | 224 | 48 | 13 | 45 | ||
-
*A full list of these sequences can be found in Table 1—source data 1.
†V Region includes all bases between the 1 st predicted nucleotide of the V segment to the last predicted nucleotide of the J segment (V and J).
-
‡V Segment includes all bases between the 1 st predicted nucleotide of the V segment to the last predicted nucleotide of the V segment (V only).
§CDR3-J includes all bases after the last predicted nucleotide of the V segment to the last predicted nucleotide of the J segment.
-
#Number of groups with identical CDR3-J sequences, which we used to determine sequence relatedness (see text for details).
**Number of groups with identical CDR3-J sequences used in this study. (Those not used contained no mutation with V segments.)
-
††Total number of sequences for each alpha V used to assess somatic hypermutation within this study (e.g., for aV1, 4 different clonal groups contained 2, 3, 3, and 5 identical CDR3-J regions, respectively).
-
Table 1–source data 1
Tab-delimited text file containing sequences used in this paper for analysis of somatic hypermutation of TcR alpha chain.
- https://doi.org/10.7554/eLife.28477.008
Frequencies of somatic hypermutation in nurse shark alpha V groups (αV G) containing the same CDR3.
Mutation frequency is the total number of nucleotide changes to a Geneious-derived consensus sequence divided by the total number of nucleotides. We counted synonymous (S) and nonsynymous (N) mutations separately for each FR and CDR for 11 different CDR3 groups in seven predicted alpha V genes. [FR: framework region; CDR: complementarity-determining region; Seqs: sequences; Nuc: nucleotides; Freq: frequency]
| FR1 | FR2 | FR3 | FR Means | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| αV Group | # Seqs | # Codons | Total | Mutations | Total | Mutation | # Codons | Total | Mutations | Total | Mutation | # Codons | Total | Mutations | Total | Mutation | ||||||
| Nuc | S | N | Mutation | Freq (%) | Nuc | S | N | Mutation | Freq (%) | Nuc | S | N | Mutation | Freq (%) | S | N | ALL | |||||
| αV1.1 | 3 | 25 | 225 | 2 | 12 | 14 | 6.222 | 17 | 153 | 0 | 2 | 2 | 1.307 | 41 | 369 | 4 | 4 | 8 | 2.168 | 6 | 18 | 24 |
| αV1.2 | 5 | 25 | 375 | 10 | 8 | 18 | 4.800 | 17 | 255 | 1 | 8 | 9 | 3.529 | 41 | 615 | 5 | 5 | 10 | 1.626 | 16 | 21 | 37 |
| αV1.4 | 3 | 25 | 225 | 1 | 0 | 1 | 0.444 | 17 | 153 | 1 | 3 | 4 | 2.614 | 41 | 369 | 4 | 5 | 9 | 2.439 | 6 | 8 | 14 |
| αV2.1 | 5 | 25 | 375 | 0 | 0 | 0 | 0.000 | 17 | 255 | 1 | 0 | 1 | 0.392 | 42 | 630 | 0 | 0 | 0 | 0.000 | 1 | 0 | 1 |
| αV3.1 | 4 | 25 | 300 | 0 | 4 | 4 | 1.333 | 17 | 204 | 0 | 0 | 0 | 0.000 | 40 | 480 | 0 | 1 | 1 | 0.208 | 0 | 5 | 5 |
| αV4.1 | 2 | 25 | 150 | 1 | 2 | 3 | 2.000 | 17 | 102 | 0 | 2 | 2 | 1.961 | 43 | 258 | 11 | 7 | 18 | 6.977 | 12 | 11 | 23 |
| αV4.2 | 2 | 25 | 150 | 0 | 0 | 0 | 0.000 | 17 | 102 | 1 | 1 | 2 | 1.961 | 43 | 258 | 2 | 7 | 9 | 3.488 | 3 | 8 | 11 |
| αV5.1 | 2 | 25 | 150 | 0 | 6 | 6 | 4.000 | 17 | 102 | 2 | 3 | 5 | 4.902 | 42 | 252 | 1 | 2 | 3 | 1.190 | 3 | 11 | 14 |
| αV7.2 | 2 | 25 | 150 | 4 | 7 | 11 | 7.333 | 17 | 102 | 1 | 3 | 4 | 3.922 | 41 | 246 | 1 | 6 | 7 | 2.846 | 6 | 16 | 22 |
| αV10.1 | 4 | 25 | 300 | 0 | 0 | 0 | 0.000 | 17 | 204 | 1 | 1 | 2 | 0.980 | 41 | 492 | 4 | 0 | 4 | 0.813 | 5 | 1 | 6 |
| αV10.2 | 9 | 25 | 675 | 2 | 11 | 13 | 1.926 | 17 | 459 | 4 | 4 | 8 | 1.743 | 40 | 1080 | 3 | 11 | 14 | 1.296 | 9 | 26 | 35 |
| Sum | 41 | 275 | 3075 | 20 | 50 | 70 | 187 | 2091 | 12 | 27 | 39 | 455 | 5049 | 35 | 48 | 83 | 67 | 125 | 192 | |||
| Mean Mutation Freq (%) | 0.65 | 1.63 | 2.28 | 0.57 | 1.29 | 1.87 | 0.69 | 0.95 | 1.64 | 0.66 | 1.22 | 1.88 | ||||||||||
| Standard Deviation | 2.994 | 4.547 | 6.531 | 1.136 | 2.252 | 2.841 | 3.125 | 3.414 | 5.429 | 4.742 | 8.201 | 11.861 | ||||||||||
| CDR1 | CDR2 | CDR3 | CDR Means | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AlphaV Group | # Seqs | # Codons | Total | Mutations | Total | Mutation | # Codons | Total | Mutations | Total | Mutation | # Codons | Total | Mutations | Total | Mutation | ||||||
| Nuc | S | N | Mutation | Freq (%) | Nuc | S | N | Mutation | Freq (%) | Nuc | S | N | Mutation | Freq (%) | S | N | ALL | |||||
| αV1.1 | 3 | 8 | 72 | 2 | 2 | 4 | 5.556 | 5 | 45 | 1 | 0 | 1 | 2.222 | 10 | 90 | 1 | 6 | 7 | 7.778 | 4 | 8 | 12 |
| αV1.2 | 5 | 8 | 120 | 5 | 4 | 9 | 7.500 | 5 | 75 | 1 | 2 | 3 | 4.000 | 8 | 120 | 1 | 4 | 5 | 4.167 | 7 | 10 | 17 |
| αV1.4 | 3 | 8 | 72 | 0 | 1 | 1 | 1.389 | 5 | 45 | 0 | 0 | 0 | 0.000 | 5 | 45 | 0 | 2 | 2 | 4.444 | 0 | 3 | 3 |
| αV2.1 | 5 | 7 | 105 | 0 | 1 | 1 | 0.952 | 5 | 75 | 0 | 0 | 0 | 0.000 | 3 | 45 | 0 | 0 | 0 | 0.000 | 0 | 1 | 1 |
| αV3.1 | 4 | 6 | 72 | 0 | 3 | 3 | 4.167 | 6 | 72 | 0 | 0 | 0 | 0.000 | 1 | 12 | 0 | 0 | 0 | 0.000 | 0 | 3 | 3 |
| αV4.1 | 2 | 7 | 42 | 3 | 1 | 4 | 9.524 | 6 | 36 | 0 | 1 | 1 | 2.778 | 6 | 36 | 2 | 0 | 2 | 5.556 | 5 | 2 | 7 |
| αV4.2 | 2 | 7 | 42 | 1 | 1 | 2 | 4.762 | 6 | 36 | 0 | 0 | 0 | 0.000 | 6 | 36 | 0 | 0 | 0 | 0.000 | 1 | 1 | 2 |
| αV5.1 | 2 | 7 | 42 | 0 | 3 | 3 | 7.143 | 6 | 36 | 0 | 7 | 7 | 19.444 | 7 | 42 | 0 | 0 | 0 | 0.000 | 0 | 10 | 10 |
| αV7.2 | 2 | 5 | 30 | 0 | 0 | 0 | 0.000 | 7 | 42 | 1 | 1 | 2 | 4.762 | 6 | 36 | 0 | 2 | 2 | 5.556 | 1 | 3 | 4 |
| αV10.1 | 4 | 7 | 84 | 3 | 0 | 3 | 3.571 | 6 | 72 | 0 | 0 | 0 | 0.000 | 7 | 84 | 0 | 3 | 3 | 3.571 | 3 | 3 | 6 |
| αV10.2 | 9 | 7 | 189 | 1 | 8 | 9 | 4.762 | 6 | 162 | 2 | 2 | 4 | 2.469 | 7 | 189 | 3 | 0 | 3 | 1.587 | 6 | 10 | 16 |
| Sum | 41 | 77 | 870 | 15 | 24 | 39 | 63 | 696 | 5 | 13 | 18 | 66 | 735 | 7 | 17 | 24 | 27 | 54 | 81 | |||
| Mean Mutation Freq (%) | 1.72 | 2.76 | 4.48 | 0.72 | 1.87 | 2.59 | 0.95 | 2.31 | 3.27 | 1.17 | 2.35 | 3.52 | ||||||||||
| Standard Deviation | 1.690 | 2.316 | 2.979 | 0.688 | 2.089 | 2.248 | 1.027 | 2.067 | 2.272 | 2.659 | 3.754 | 5.626 | ||||||||||
Number and frequency of DNA mutations that occur in tandem within framework regions (FR) and complementarity determining regions (CDR) in nurse shark alpha V (αV) groups.
All mutations include both tandem and point mutations within a region. [Seqs: sequences]
| FR | CDR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # Nucleotides Tandemly Mutated | All mutation | Frequency of tandem mutation | # Nucleotides Tandemly Mutated | All mutation | Frequency of tandem mutation | ||||||||
| αV Group | # Seqs | 2 | 3 | 4 | Sum | 2 | 3 | 4 | Sum | ||||
| αV1.1 | 3 | 1 | 0 | 1 | 6 | 24 | 25.0 | 3 | 0 | 0 | 6 | 12 | 50.0 |
| αV1.2 | 5 | 4 | 1 | 0 | 11 | 37 | 29.7 | 5 | 1 | 0 | 13 | 17 | 76.5 |
| αV1.4 | 3 | 3 | 0 | 0 | 6 | 14 | 42.9 | 0 | 0 | 0 | 0 | 3 | 0.0 |
| αV2 | 5 | 0 | 0 | 0 | 0 | 1 | 0.0 | 0 | 0 | 0 | 0 | 1 | 0.0 |
| αV3 | 4 | 2 | 0 | 0 | 4 | 5 | 80.0 | 1 | 0 | 0 | 2 | 3 | 66.7 |
| αV4.1 | 2 | 4 | 1 | 0 | 11 | 23 | 47.8 | 0 | 1 | 0 | 3 | 7 | 42.9 |
| αV4.2 | 2 | 1 | 1 | 0 | 5 | 11 | 45.5 | 0 | 1 | 0 | 3 | 2 | 150.0 |
| αV5 | 2 | 1 | 0 | 0 | 2 | 14 | 14.3 | 1 | 1 | 0 | 5 | 10 | 50.0 |
| αV7.2 | 2 | 4 | 1 | 0 | 11 | 22 | 50.0 | 1 | 0 | 0 | 2 | 4 | 50.0 |
| αV10.1 | 4 | 3 | 0 | 0 | 6 | 6 | 100.0 | 0 | 0 | 0 | 0 | 6 | 0.0 |
| αV10.2 | 9 | 4 | 1 | 0 | 11 | 35 | 31.4 | 2 | 1 | 0 | 7 | 16 | 43.8 |
| Total | 41 | 27 | 5 | 1 | 73 | 192 | 38.0 | 13 | 5 | 0 | 41 | 81 | 50.6 |
Target nucleotide mutation frequency in DGYW/WRCH or WA/TW mutation hotspots within framework regions (FR) and complementarity determining regions (CDR).
[DGYW/WRCH (G:C is the mutable position; D = A/G/T, Y = C/T, W = A/T, R = A/G, and H = T/C/A); WA/TW (A:T is the mutable position; W = A/T); ‘ALL’ refers to nucleotides within a hotspot motif; ‘All other’ refers to nucleotides outside a hotspot motif]
| Mutated base | Hotspot motif | Region | Total nucleotides | Observed # Mutations | Expected # Mutations | Mutation freq (%) | χ2 p | T-test P |
|---|---|---|---|---|---|---|---|---|
| G/C | DGYW/WRCH | FR | 927 | 56 | 18.16 | 6.04 | 0.0000 | 0.0267 |
| CDR | 400 | 35 | 26.22 | 8.75 | ||||
| ALL | 1327 | 91 | 37.52 | 6.86 | 0.0000 | |||
| Outside Motif | 4402 | 71 | 124.48 | 1.61 | ||||
| A/T | WA/TW | FR | 2531 | 54 | 36.07 | 2.13 | 0.0015 | 0.2248 |
| CDR | 400 | 26 | 21.12 | 4.03 | ||||
| ALL | 2931 | 80 | 55.97 | 2.52 | 0.0000 | |||
| Outside Motif | 3690 | 41 | 65.03 | 1.11 | ||||
-
*T-test analysis was used to compare mutations within hotspot motifs to those outside hotspot motifs. Mutations to G and C nucleotides occurred significantly more often within DGYW/WRCH motifs than outside these motifs, while mutations to A and T nucleotides showed no preference for WA/TW motifs.
†χ2 analysis was used to compare observed and expected numbers of mutations between FR and CDR regions and between mutations inside and outside hotspot motifs. More mutations to all nucleotides occurred within hotspots than outside hotspots, and significantly more mutations occurred to nucleotides within CDRs than FRs.
Bias in base substitution during somatic hypermutation of TcR alpha V genes within all sequence regions (ALL), framework regions (FR), or complementarity determining regions (CDR).
Probability of occurrence is the proportion of that base out of the total nucleotides. [Nuc: nucleotides; OBS: Observed; EXP: expected; MI: mutability index; ChiSq: Chi squared]
| a | ALL | Base | Occurrence | Probability of occurrence | OBS | EXP | MI* | ChiSq |
|---|---|---|---|---|---|---|---|---|
| G | 2895 | 0.230 | 77 | 65.05 | 1.18 | 0.0022 | ||
| C | 2834 | 0.225 | 85 | 63.68 | 1.33 | |||
| A | 3498 | 0.278 | 64 | 78.60 | 0.81 | 0.0068 | ||
| T | 3368 | 0.267 | 57 | 75.68 | 0.75 | |||
| Total | 12595 | 1.00 | 283 | 283 | ||||
| GC Mutation: 57.0%; Transitions: 42.8%; Transversions: 25.1% | ||||||||
| b | FR | Base | Occurrence | Probability of Occurrence | OBS | EXP | MI* | ChiSq |
| G | 2378 | 0.23 | 58 | 44.50 | 1.30 | 0.0037 | ||
| C | 2268 | 0.22 | 56 | 42.44 | 1.32 | |||
| A | 2779 | 0.27 | 38 | 52.00 | 0.73 | 0.0082 | ||
| T | 2835 | 0.28 | 40 | 53.05 | 0.75 | |||
| Total | 10260 | 1.00 | 192 | 192 | ||||
| GC Mutation: 59.0%; Transitions: 41.7%; Transversions: 24.0% | ||||||||
| c | CDR | Base | Occurrence | Probability of Occurrence | OBS | EXP | MI | ChiSq |
| G | 517 | 0.22 | 19 | 20.15 | 0.94 | 0.1336 | ||
| C | 566 | 0.24 | 29 | 22.06 | 1.31 | |||
| A | 719 | 0.31 | 26 | 28.02 | 0.93 | 0.3620 | ||
| T | 533 | 0.23 | 17 | 20.77 | 0.82 | |||
| Total | 2335 | 1.00 | 91 | 91 | ||||
| GC Mutation: 52.9%; Transitions: 45.1%; Transversions: 27.5% | ||||||||
-
*Mutability Index, as first defined in Chen et al., 2012. χ2 analysis was used to compare observed and expected numbers of mutations. G and C mutated significantly more often than expected, while A and T mutated significantly less often than expected. Base composition: 23.0% G, 22.5% C 27.8% A, 26.7% T.
Frequencies of somatic hypermutation in nurse shark thymus and peripheral lymphoid tissue (blood and spiral valve).
Mutations were analyzed only in alpha V groups containing the same third complementarity-determining region (CDR). Mutation frequency was measured as the total number of nucleotide changes to a Geneious-derived consensus sequence divided by the total number of nucleotides in all sequences. Nonsynymous (N) and synonymous (S) mutations (mut) were counted separately for each framework (FR) and CDR for two predicted alpha V genes. [FR1, FR2, FR3, CDR1, CDR2, and CDR3 refer to the first, second, or third FR or CDR region, respectively.]
| Tissue type | Mut type | FR Mutations (#) | CDR Mutations (#) | FR mutation frequency | CDR mutation frequency | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FR1 | FR2 | FR3 | All FR | CDR1 | CDR2 | CDR3 | All CDR | FR1 | FR2 | FR3 | CDR1 | CDR2 | CDR3 | ||
| Thymus (6 sequences) | N | 8 | 8 | 7 | 23 | 1 | 3 | 3 | 7 | 0.570 | 0.871 | 0.317 | 0.071 | 0.327 | 0.136 |
| S | 8 | 2 | 6 | 16 | 1 | 0 | 0 | 1 | 0.570 | 0.218 | 0.272 | 0.071 | 0.000 | 0.000 | |
| ALL | 16 | 10 | 13 | 39 | 2 | 3 | 3 | 8 | 1.140 | 1.089 | 0.590 | 0.529 | 1.075 | 1.235 | |
| Total Nucleotides | 1404 | 918 | 2205 | 4527 | 378 | 279 | 243 | 900 | |||||||
| Periphery (2 sequences) | N | 7 | 4 | 6 | 17 | 0 | 2 | 2 | 4 | 1.496 | 1.307 | 0.833 | 0.000 | 1.852 | 1.587 |
| S | 4 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 0.855 | 0.000 | 0.139 | 0.000 | 0.000 | 0.000 | |
| ALL | 11 | 4 | 7 | 22 | 0 | 2 | 2 | 4 | 2.350 | 1.307 | 0.972 | 0.000 | 1.852 | 1.587 | |
| Total Nucleotides | 468 | 306 | 720 | 1494 | 126 | 108 | 126 | 360 | |||||||
List of forward (F) and reverse (R) primers used to generate T cell receptor (TcR) sequences and expression data.
[AID: Activation induced cytidine deaminase; B2M: beta-2 microglobulin; α: alpha; β: beta; γ: gamma; δ: delta; V: variable region; C: constant region].
| Primer | F/R | ID | Location | Nucleotide sequence (5' to 3') | Amino acid | Tm |
|---|---|---|---|---|---|---|
| TcR αV1 | F | MFC370 | leader region of αV1 | ATG TTG CCT GAA GCT C | MLPEA | 55 |
| R | MFC191 | alpha C region | CAT TGG TGG ATA GCA AGC CCT TCG AT | SKGLLSTN | 76 | |
| TcR αV4 | F | MFC122 | beginning of αV4 | GTC TCC TCA GTT GTT CGT AC | VSSVVR | 58 |
| R | MFC123 | end of αV4 | CAG TAA TAC ACA GCA GCG TC | DAAVYY | 58 | |
| F | MFC374 | leader region of αV4 | TGG ATT GTG TGG GCA GTA | WIVWAV | 54 | |
| R | MFC191 | alpha C region | CAT TGG TGG ATA GCA AGC CCT TCG AT | SKGLLSTN | 76 | |
| TcR αV5 | F | MFC124 | beginning of αV5 | CTC AGG AAG GAG AGA TTA TCA C | QEGEII | 60 |
| R | MFC125 | end of αV5 | CAA TGA TAC ACG GCG GAG TC | DSAVYH | 60 | |
| F | MFC124 | beginning of αV5 | CTC AGG AAG GAG AGA TTA TCA C | QEGEII | 60 | |
| R | MFC191 | alpha C region | CAT TGG TGG ATA GCA AGC CCT TCG AT | SKGLLSTN | 76 | |
| TcR αV7 | F | MFC376 | end of leader αV7 | AGC GAT GGA GTT TCT GTG ATT | SDGVSVI | 58 |
| R | MFC191 | alpha C region | CAT TGG TGG ATA GCA AGC CCT TCG AT | SKGLLSTN | 76 | |
| TcR αV10 | F | MFC378 | leader region of αV10 | CTA TTT CTT CAC TAC CGC AG | YFFTTA | 56 |
| R | MFC191 | alpha C region | CAT TGG TGG ATA GCA AGC CCT TCG AT | SKGLLSTN | 76 | |
| TcR α 5' | F | GeneRacer 5' Nested | homologous to RNA oligo | GGA CAC TGA CAT GGA CTG AAG GAG TA | -- | 78 |
| TcR α 3' | R | MFC191 | alpha C region | CAT TGG TGG ATA GCA AGC CCT TCG AT | SKGLLSTN | 76 |
| TcR βV1 | F | MFC126 | beginning of bV1 | CTC CGT ACA TCG TCT CTA TTG | PYIVSI | 60 |
| R | MFC127 | end of βV1 | CAC GCA CAG AAA TAG ACA GC | AVYFCA | 58 | |
| TcR βV2 | F | MFC128 | beginning of βV2 | CTA CGT GGA GCA GTC TCC ATC | YVEQSP | 63 |
| R | MFC129 | end of βV2 | GCA CGC ACA ATA ATA GAC AGC C | AVYYCAC | 62 | |
| TcR βV3 | F | MFC130 | beginning of βV3 | CTA CGT GGA ACA GTC TCC TTC | YVEQSP | 61 |
| R | MFC131 | end of βV3 | CAC GCG CAG AAA TAG ACA G | VYFCA | 57 | |
| TcR βV5 | F | MFCb50 | beginning of βV5 | GTT CGG TGC TCT TTC TCT GC | MFGALSLH | 60 |
| R | MFCb54 | end of βV5 | GAC TGC AGT ATC AGT CGG CAC C | LVPTDTAV | 66 | |
| TcR γV1 | F | MFCg56 | beginning of γV1 | GTC GCT GTA TTA CTG GCT CAT TG | MSLYYWL | 63 |
| R | MFCg59 | end of γV1 | GAG CGC ACA GTA ATA GGT GGC AG | TATYYCAL | 67 | |
| TcR γV3 | F | MFCg58 | beginning of γV3 | GAA GGG TCA CGT CCT TGC G | MKGHVLA | 62 |
| R | MFCg61 | end of γV3 | GAT CCC AGA GTC ATC CTC | EDDSGI | 56 | |
| F | MFC170 | beginning of γV3 | CAA TAA CCA GAG CAC CGG G | ITRAP | 56 | |
| R | MFC171 | end of γV3 | AGA TCC CAG AGT CGT CCT C | EDDSGI | 56 | |
| TcR δV3 | F | MFCd62 | beginning of δV3 | GAT TCC CCG TCC CTG GTG TC | DSPSLVS | 65 |
| R | MFCd66 | end of δV3 | CAG TGC ACA GTG ATA CAC AGC | AVYHCAL | 61 | |
| TcR δV5 | F | MFCd63 | beginning of δV5 | GCA GCT ACT CAG TAT CTG G | MQLLSIW | 57 |
| R | MFCd67 | end of δV5 | GAA AGC ACA GTA ATA CAG AG | ALYYCAF | 54 | |
| TcR δV7 | F | MFC172 | beginning of δV7 | CTG TCA CTC AGT TAT TCT CCT C | VTQLFS | 60 |
| R | MFC173 | end of δV7 | GCA GCC CAG TTA TAG TCA AAC | LTITGL | 60 | |
| TcR δV12 | F | MFC174 | beginning of δV12 | CAG AGC CCA CCT CAG TTA C | QSPPQ; | 60 |
| R | MFC175 | end of δV12 | GAG CGC AGT AAT AGA TGG C | AIYYCA | 57 | |
| TcR δV16 | R | MFC176 | end of δV16 | GCA GCT CCG AGA TAG ACA AC | LSISEL | 60 |
| F | MFC177 | beginning of δV16 | GAG TCC TGG CTC ACG CAA TC | ESWLTQ | 63 | |
| TcR δV17 | F | MFC178 | beginning of δV17 | CAG TCT TGG TCA GAA ATA ACC | QSWSEIT | 57 |
| R | MFC179 | end of δV17 | CAA CTG AAG ATA AGT GAT CG | ITYLQL | 54 | |
| AID | F | MFC342 | beginning of AID exon 1 | AGG CAC GAG ACC TAC ATG TTG | RHETYML | 61 |
| R | MFC347 | end of AID exon 2 | TGA ACC AGG TGA GGC GGT A | YRLTWF | 60 | |
| B2M | F | MFC211 | first cysteine | AAC GTG TTG CTC TGT CAT GC | NVLLCHA | 58 |
| R | MFC212 | before second cysteine | GGG GTG AAC TCC ACA TAA CG | RYVEFTP | 60 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28477.026





