Prolyl dihydroxylation of unassembled uS12/Rps23 regulates fungal hypoxic adaptation
Figures
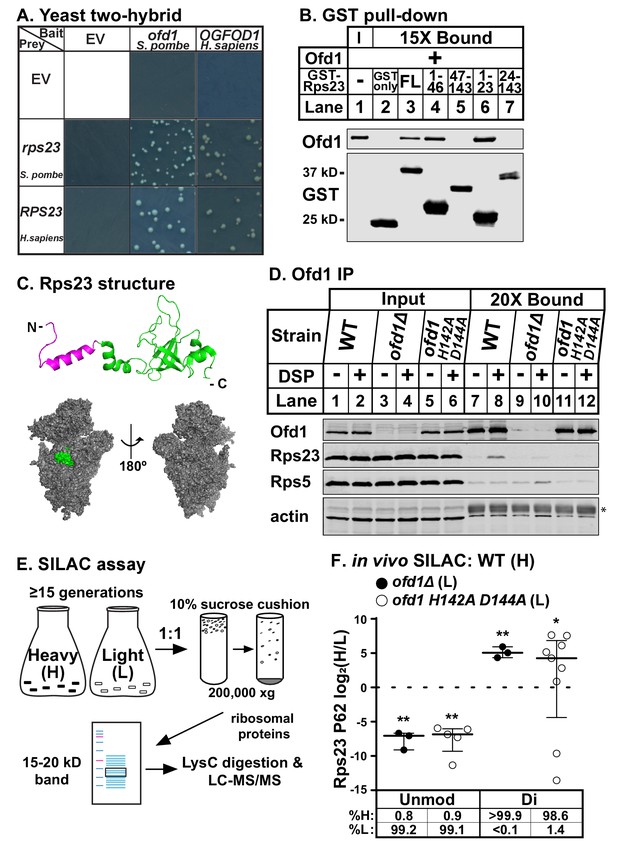
Ofd1 binds and dihydroxylates Rps23 P62.
(A) Yeast two-hybrid assays were performed using S. cerevisiae AH109 strain transformed with indicated bait and prey vectors, either empty vector (EV) or expressing specified genes. Cells were grown on SD-Leu-Ade-Trp-His medium supplemented with X-α-Gal for 8 days. Shown are cropped regions of plates; see Figure 1—figure supplement 1A for uncropped plates. (B) Full-length (FL) and truncated Rps23 were N-terminally tagged with GST and purified on glutathione beads from bacterial lysates. Following incubation with purified Ofd1 (84.5 nM), bound proteins were eluted with reduced glutathione. Input and bound fractions were analyzed by immunoblot using antibodies against Ofd1 and GST. Shown are representative blots from one of three independent experiments. (C) Top: crystal structure of Rps23 in the S. cerevisiae ribosome (PDB 4V88) with the Ofd1-binding site, Rps23 (aa 1–23), colored magenta; bottom: surface representation of Rps23 in the 40S subunit (left: interface; right: solvent-exposed). (D) Wild-type, ofd1Δ, and ofd1 H142A D144A cells were treated with vehicle (8% DMSO) or crosslinker (2 mM DSP) for 5 min, lysed in detergent, and centrifuged to pellet ribosomes. Ofd1 was purified from ribosome-depleted lysates with anti-Ofd1 antibody, and input and bound fractions were analyzed by immunoblot with indicated antibodies. Asterisk (*) denotes IgG heavy chain (see Figure 1—figure supplements 1–2 for additional information). Shown are representative blots from one of three independent experiments. (E) Diagram outlining SILAC assay to quantify Rps23 hydroxylation. Yeast cells were cultured in SILAC medium supplemented with either heavy (H) or light (L) lysine for ≥ 15 generations. Whole cell lysates were mixed 1:1 H:L and centrifuged through a sucrose cushion to collect ribosomes. Ribosomal proteins were extracted from pellets and separated by gel electrophoresis prior to LysC digestion and LC-MS/MS. (F) Wild-type, ofd1Δ, and ofd1 H142A D144A cells were cultured and processed as described in (E) with wild-type cells labeled with heavy lysine. Error bars represent the interquartile range of H/L ratios containing either unmodified (unmod) or dihydroxylated (di) Rps23 P62, and significance was tested by Mann-Whitney (*p<0.01; **p<0.0001; n.s.). Median %H and %L values are reported below. Shown are PSMs from one of four independent experiments. See also Figure 1—source data 1.
-
Figure 1—source data 1
SILAC mass spectrometry data.
SILAC ratios used to quantify Rps23 P62 hydroxylation shown in Figure 1F and Figure 4A–B.
- https://doi.org/10.7554/eLife.28563.006
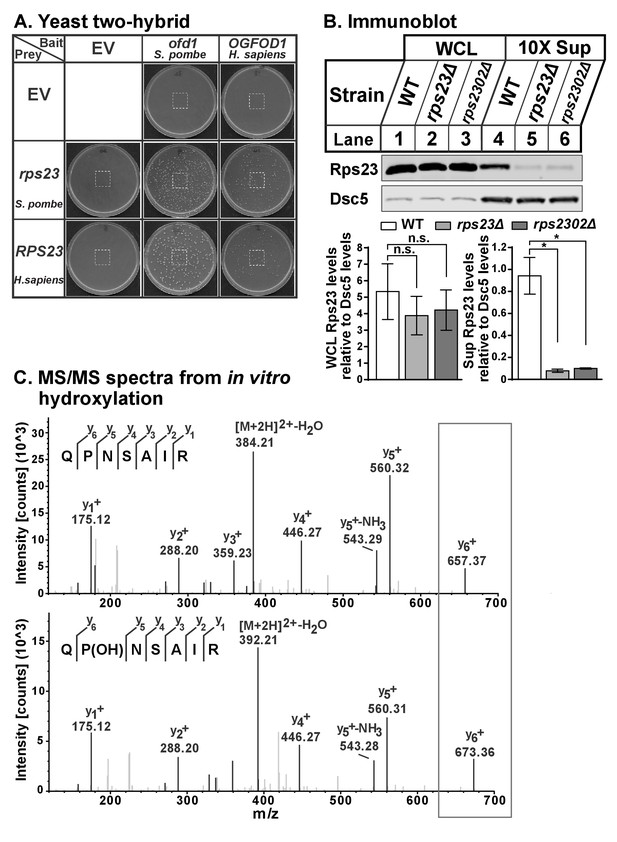
Ofd1 binds and dihydroxylates Rps23 P62.
(A) Yeast two-hybrid assays were performed using S. cerevisiae AH109 strain transformed with indicated bait and prey vectors, either EV or expressing specified genes. Cells were grown on SD-Leu-Ade-Trp-His medium supplemented with X-α-Gal for 8 days. Cropped regions of plates used in Figure 1 are indicated. (B) Wild-type, rps23Δ, and rps2302Δ cells were cultured in rich medium, lysed in detergent, and centrifuged to pellet ribosomes. Whole cell extracts and ribosome-cleared supernatant (sup) were analyzed by immunoblot with antibodies against Rps23 and Dsc5. Shown below representative blots are mean ± SEM of three independent experiments; significance was determined by paired, one-tailed t test (*p<0.05; n.s., not significant). (C) In vitro hydroxylation reaction containing RPS23 and Ofd1 was incubated overnight with 2OG, Fe(II), ascorbate, and BSA, then digested with trypsin for 2 hr followed by LC-MS/MS. Shown are the y ion series for RPS23 QPNSAIR peptide: unmodified (top) and monohydroxylated P62 (bottom).
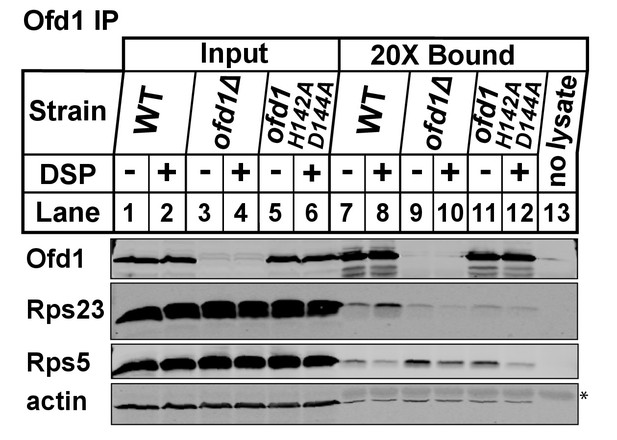
Wild-type, ofd1Δ, and ofd1 H142A D144A cells were treated and processed as described in Figure 1D.
Ofd1 was purified from ribosome-depleted lysates using anti-Ofd1 antibody. A no lysate control (lane 13) was included to identify signals from the anti-Ofd1 antibody alone. Input and bound fractions were analyzed by immunoblot with indicated antibodies. Asterisk (*) denotes IgG heavy chain.
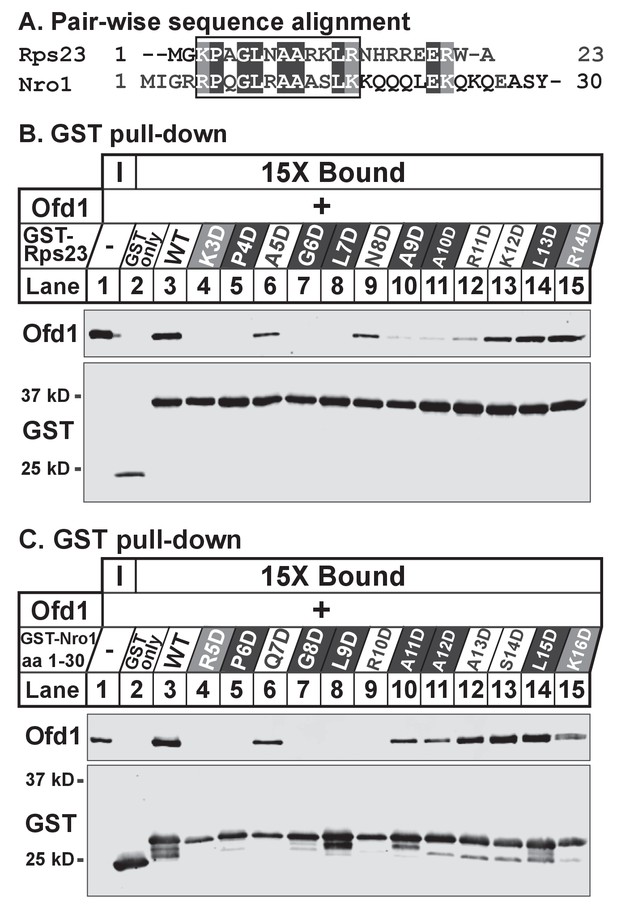
Identification of an Ofd1 binding sequence.
(A) Pair-wise alignment of Rps23 (aa 1–23) and Nro1 (aa 1–30) sequences required to bind Ofd1; black boxes denote identical residues and gray boxes denote similar residues. Amino acids enclosed by the box were individually mutated and tested for binding to Ofd1. (B, C) Lysates of bacteria expressing wild-type or mutant GST-Rps23 FL (B) or GST-Nro1 aa 1–30 (C) were incubated with glutathione beads prior to incubation with purified Ofd1 (84.5 nM). Input and bound fractions were analyzed by immunoblot using antibodies against Ofd1 and GST. Shown are representative blots from one of at least two independent experiments.
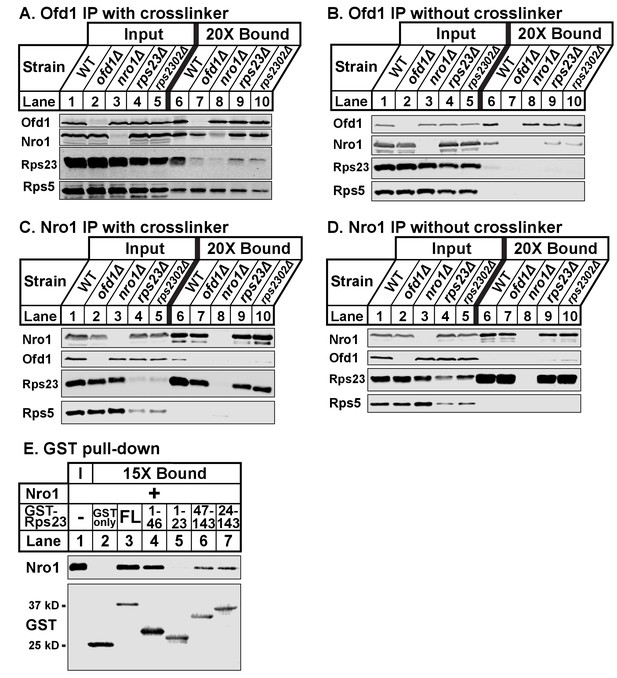
Ofd1, Rps23, and Nro1 form a complex.
(A, B) Wild-type, ofd1Δ, nro1Δ, rps23Δ, and rps2302Δ cells were treated with crosslinker (A) (2 mM DSP) or without crosslinker (B), lysed in detergent, and depleted of ribosomes. Ofd1 was purified from lysates using anti-Ofd1 antibody, and input and bound fractions were analyzed by immunoblot with indicated antibodies. (C, D) Wild-type, nro1Δ, ofd1Δ, rps23Δ, and rps2302Δ cells were treated in the presence of crosslinker (C) or absence (D), lysed in detergent, and depleted of ribosomes. Nro1 was isolated from lysates using anti-Nro1 antibody, and input and bound fractions were analyzed by immunoblot with indicated antibodies. (E) GST-tagged full length (FL) and truncated Rps23 were purified on glutathione beads and incubated with purified Nro1 (84.5 nM). Input and bound fractions were analyzed by immunoblot using antibodies against Nro1 and GST. Representative blots from one of two independent experiments are shown for all figures.
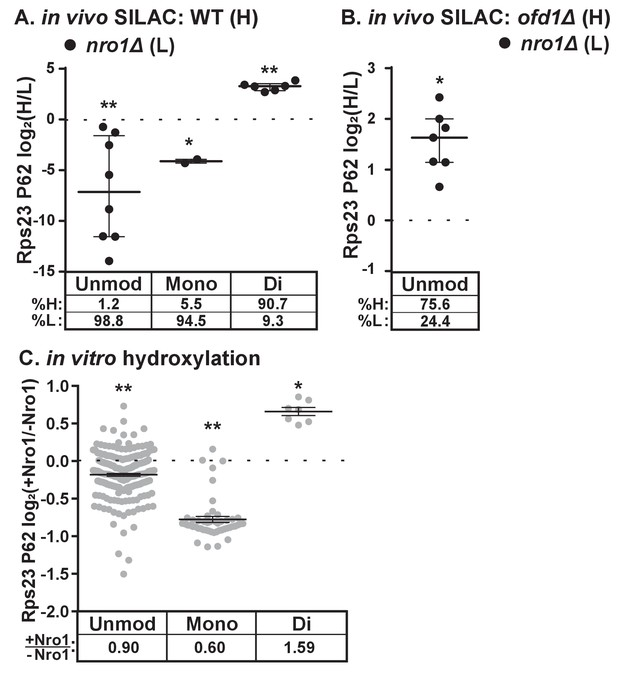
Dihydroxylation of Rps23 P62 requires Nro1.
(A) Wild-type and nro1Δ cells were cultured and processed as described in Figure 1E with wild-type cells labeled with heavy lysine. Error bars represent the interquartile range and significance was determined by Mann-Whitney (*p<0.001, **p<0.0001). Median % heavy and % light are reported below. Shown are PSMs from one of four independent experiments. (B) ofd1Δ and nro1Δ cells were cultured and processed as described in Figure 1E with ofd1Δ cells labeled with heavy lysine to determine the relative amount of unmodified Rps23 P62 in nro1Δ cells. Mann-Whitney test, *p<0.0001. Median % heavy and % light are reported below. Shown are PSMs from one of two independent experiments. See also Figure 1—source data 1. (C) Ofd1 (0.5 μM) and MBP-Rps23 (5 μM) were incubated in the absence and presence of Nro1 (5 μM) as described in Methods for 1 hr followed by quantification of Rps23 P62 hydroxylation by TMT LC-MS/MS. Error bars represent ± SEM. Significance was calculated by Wilcoxon signed rank test (*p<0.05; **p<0.0001). Average fold changes (+Nro1/-Nro1) are reported below. Shown are PSMs from one of two independent experiments. See also Figure 4—source data 1.
-
Figure 4—source data 1
TMT mass spectrometry data.
TMT data used to quantify Rps23 P62 hydroxylation in vitro shown in Figure 4C.
- https://doi.org/10.7554/eLife.28563.010
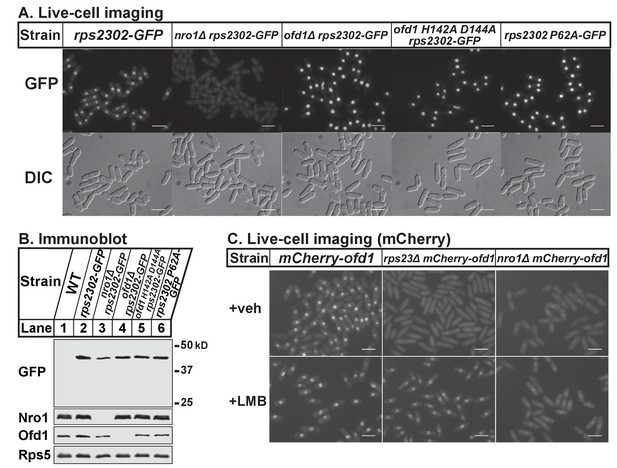
Nro1 imports Rps23.
(A) Indicated strains were cultured in rich medium and imaged by fluorescence and differential interference contrast (DIC) microscopy. Scale bar = 10 microns. Shown are representative images from one of at least two independent experiments. (B) Whole cell extracts (50 μg) of indicated strains were analyzed by immunoblot with antibodies against GFP, Nro1, Ofd1, and Rps5. Shown are representative blots from one of at least two independent experiments. (C) mCherry-ofd1, rps23Δ mCherry-ofd1, and nro1Δ mCherry-ofd1 cells were treated with vehicle (0.05% [v/v] ethanol) or the nuclear export inhibitor LMB (92.5 nM) for 1 hr prior to live-cell imaging by fluorescence microscopy. Scale bar = 10 microns. Shown are representative images from one of three independent experiments.
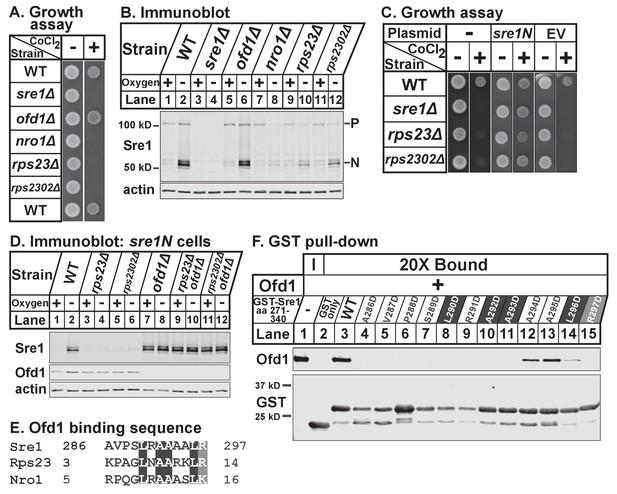
Unassembled Rps23 regulates Ofd1 inhibition of Sre1N.
(A) Indicated strains were grown on rich medium in the absence and presence of 1.6 mM cobalt chloride and imaged after 3 and 4 days, respectively (5000 cells per spot). Shown are representative images from one of two independent experiments. (B) Indicated strains were cultured in rich medium in the presence or absence of oxygen for 3 hr. Whole cell extracts (20 μg) were treated with alkaline phosphatase and analyzed by immunoblot with antibodies against Sre1 and actin. P=precursor; N=nuclear. Shown are representative blots from one of two independent experiments. (C) Wild-type, sre1Δ, rps23Δ, and rps2302Δ cells were transformed with EV or CaMV-sre1N expressing vector and grown on rich medium for 3 days or rich medium containing 1.6 mM cobalt chloride (CoCl2) for 6 days (5000 cells per spot). Shown are representative images from one of two independent experiments. (D) Indicated sre1N strains were cultured in rich medium in the presence or absence of oxygen for 3 hr. Whole cell extracts (20 μg) were phosphatase-treated and analyzed by immunoblot with antibodies against Sre1, Ofd1, and actin. Shown are representative blots from one of two independent experiments. (E) Multiple sequence alignment of Sre1 aa 286–297, Rps23 aa 3–14, and Nro1 aa 5–16. (F) Lysates from bacteria expressing wild-type or mutant GST-Sre1 aa 271–340 were incubated with glutathione beads and then with purified Ofd1 (84.5 nM). Input and bound fractions were analyzed by immunoblot using antibodies against Ofd1 and GST. Shown are representative blots from one of two independent experiments.
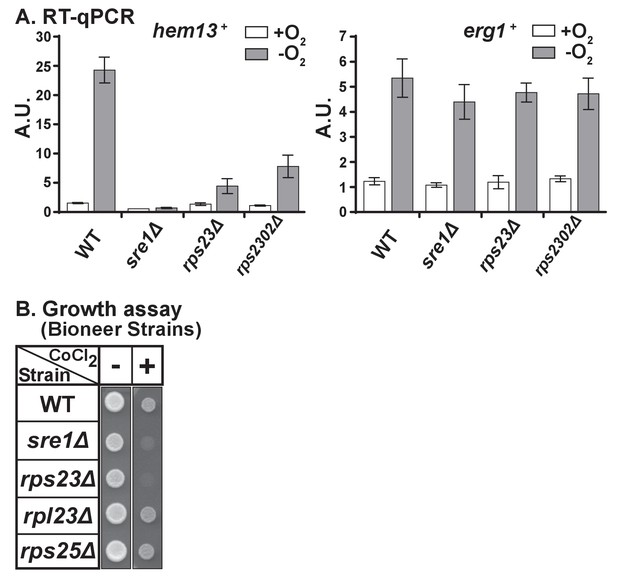
Unassembled Rps23 regulates Ofd1 inhibition of Sre1N.
(A) RNA was harvested from wild-type, sre1Δ, rps23Δ, and rps2302Δ cells grown in the presence or absence of oxygen for 3 hr. Extracted total RNA was analyzed by RT-qPCR for expression of Sre1 target hem13+ (left) and non-Sre1 target erg1+ (right). Error bars represent ± SEM of fold changes from three biological replicates with each biological replicate consisting of two technical replicates. Each biological replicate was an independent experiment. (B) Indicated strains from Bioneer haploid deletion collection were grown (5000 cells/spot) on rich medium (3 days) and rich medium containing 1.6 mM cobalt chloride (CoCl2) (6 days). Shown are representative images from one of two independent experiments.
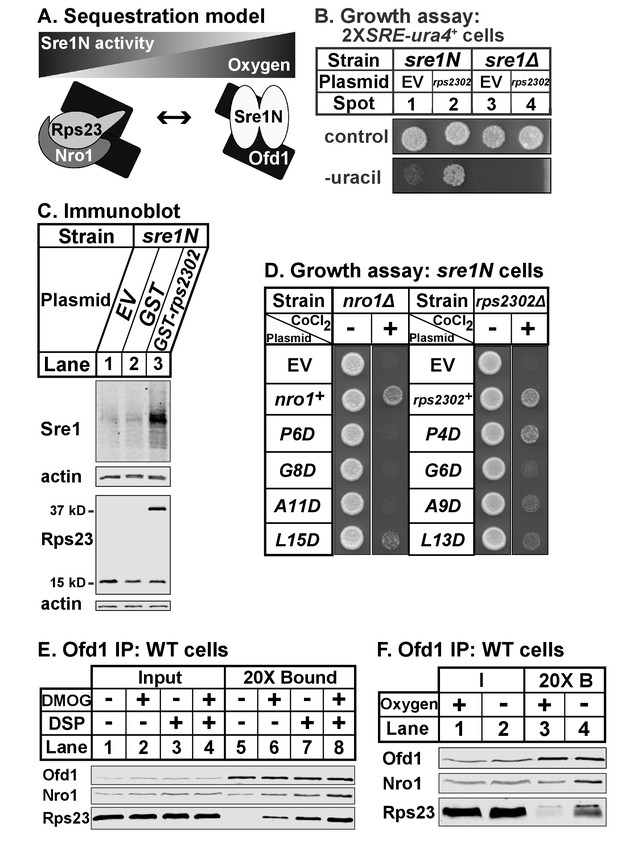
Ofd1-Rps23-Nro1 complex sequesters Ofd1 under hypoxia to activate Sre1N.
(A) Sequestration model showing Ofd1 bound to either Rps23-Nro1 or Sre1N dimer. (B) sre1N or sre1Δ cells carrying an integrated 2XSRE-ura4+ reporter gene and transformed with either EV or rps2302+ under control of the adh1 promoter were grown on EMM-Leu or EMM-Leu-Ura media (1000 cells per spot). Shown are representative images from one of two independent experiments. (C) sre1N cells carrying either EV or plasmids overexpressing GST or GST-rps2302 from the nmt1 promoter were grown in minimal medium lacking thiamine for 20 hr. Whole cell extracts (100 μg) were analyzed by immunoblot using antibodies against Sre1, Rps23, and actin. Shown are representative blots from one of three independent experiments. (D) nro1Δ sre1N and rps2302Δ sre1N cells transformed with indicated plasmids under control of the CaMV and adh promoters, respectively, were grown (5000 cells/spot) on rich medium (3 days) and rich medium supplemented with 1.6 mM CoCl2 (4 days). Shown are representative images from one of two independent experiments. (E) Wild-type cells were cultured in rich medium in the presence of DMOG (20 mM) or DMSO (1% [v/v]) for 3 hr prior to treatment with vehicle or crosslinker. Samples were lysed in detergent, depleted of ribosomes, and incubated with anti-Ofd1 antibodies. Input and bound fractions were analyzed by immunoblot with indicated antibodies. Shown are representative blots from one of two independent experiments. (F) Wild-type cells were cultured in rich medium in the presence or absence of oxygen for 3 hr prior to treatment with crosslinker then processed as in (E). Shown are representative blots from one of two independent experiments.
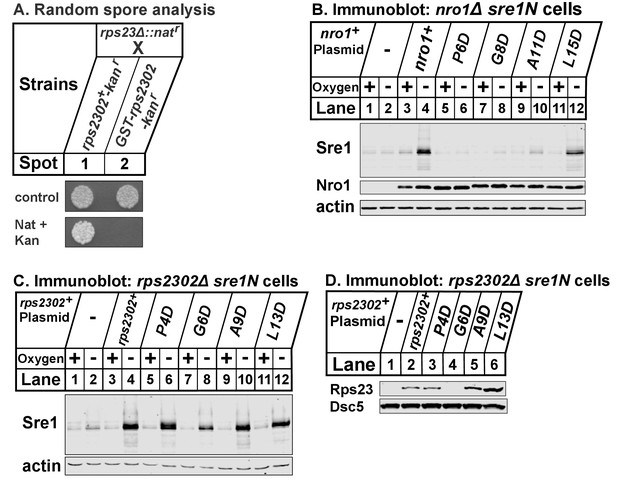
(A) rps23Δ::natr cells were mated to rps2302+-kanr and GST-rps2302-kanr cells.
Cells were incubated with glusulase to select for meiotic products. Spores were plated and grown on rich medium for 2 days (control) or rich medium supplemented with nourseothricin (nat) and kanamycin (kan) for 8 days to select for either rps2302+-kanr rps23Δ::natr or GST-rps2302-kanr rps23Δ::natr cells. Shown are representative images from one of two independent experiments. (B) nro1Δ sre1N cells transformed with the indicated plasmids under control of the CaMV promoter were grown in rich medium in the presence or absence of oxygen for 3 hr. Whole cell extracts (20 μg) were phosphatase-treated and analyzed by immunoblot using antibodies against Sre1, Nro1, and actin. Shown are representative blots from one of two independent experiments. (C) rps2302Δ sre1N cells carrying the indicated plasmids under control of the adh promoter were cultured and processed as described in (B) and analyzed using the indicated antibodies. Shown are representative blots from one of two independent experiments. (D) Transformed rps2302Δ sre1N cells from (C) were cultured in rich medium for 3 hr, lysed in detergent, and centrifuged to pellet ribosomes. Ribosome-cleared supernatant (sup) were analyzed by immunoblot with antibodies against Rps23 and Dsc5. Shown are blots from one experiment.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (S. pombe) | WT | ATCC | KGY425: h- his3-D1 leu1-32 ura4-D18 ade6-M210 | |
| strain, strain background (S. pombe) | ofd1Δ | this study | PEY1801: h- his3-D1 leu1-32 ura4-D18 ade6-M210 ofd1Δ::natMX6 | |
| strain, strain background (S. pombe) | ofd1 H142A D144A | PMID: 18418381 | PEY1152: h- his3-D1 leu1-32 ura4-D18 ade6-M210 ofd1 H142A D144A | |
| strain, strain background (S. pombe) | nro1Δ | this study | PEY1802: h- his3-D1 leu1-32 ura4-D18 ade6-M210 nro1Δ::natMX6 | |
| strain, strain background (S. pombe) | rps2302Δ | this study | PEY1803: h+ his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302Δ::natMX6 | |
| strain, strain background (S. pombe) | rps23Δ; rps23Δ-natr | this study | PEY1804: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps23Δ::natMX6 | |
| strain, strain background (S. pombe) | rps2302-GFP | this study | PEY1805: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302+- GFP(S65T)-kanMX6 | |
| strain, strain background (S. pombe) | rps2302-GFP nro1Δ | this study | PEY1806: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302+- GFP(S65T)-kanMX6 nro1Δ::natMX6 | |
| strain, strain background (S. pombe) | rps2302-GFP ofd1Δ | this study | PEY1847: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302+- GFP(S65T)-kanMX6 ofd1Δ::natMX6 | |
| strain, strain background (S. pombe) | rps2302-GFP ofd1 H142A D144A | this study | PEY1848: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302+- GFP(S65T)-kanMX6 ofd1 H142A D144A | |
| strain, strain background (S. pombe) | rps2302 P62A-GFP | this study | PEY1849: h+ his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302 P62A-GFP(S65T)- TADH1- PURA4-kanr-TTEF | |
| strain, strain background (S. pombe) | mCherry-ofd1 | this study | PEY1807: h- his3-D1 leu1-32 ura4-D18 ade6-M210 mCherry- ofd1-TADH1-PURA4-kanr-TTEF | |
| strain, strain background (S. pombe) | mCherry-ofd1 rps23Δ | this study | PEY1808: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps23Δ:: natMX6 mCherry-ofd1-TADH1- PURA4-kanr-TTEF | |
| strain, strain background (S. pombe) | mCherry-ofd1 nro1Δ | this study | PEY1809: h- his3-D1 leu1-32 ura4-D18 ade6-M210 nro1Δ::kanMX6 mCherry-ofd1-TADH1- PURA4-kanr-TTEF | |
| strain, strain background (S. pombe) | rps2302Δ sre1N | this study | PEY1811: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302Δ:: natMX6 sre1(aa1-440) | |
| strain, strain background (S. pombe) | rps23Δ sre1N | this study | PEY1812: h- his3-D1 leu1-32 ura4-D18 ade6-M210 rps23Δ:: natMX6 sre1(aa1-440) | |
| strain, strain background (S. pombe) | rps23Δ ofd1Δ sre1N | this study | PEY1813: h+ his3-D1 leu1-32 ura4-D18 ade6-M210 ofd1Δ::kanMX6 rps23Δ:: natMX6 sre1(aa1-440) | |
| strain, strain background (S. pombe) | rps2302Δ ofd1Δ sre1N | this study | PEY1814: h+ his3-D1 leu1-32 ura4-D18 ade6-M210 ofd1Δ::kanMX6 rps2302Δ::natMX6 sre1(aa1-440) | |
| strain, strain background (S. pombe) | sre1Δ | PMID: 15797383 | PEY522: h- his3-D1 leu1-32 ura4-D18 ade6-M210 sre1Δ::kanMX6 | |
| strain, strain background (S. pombe) | WT | Bioneer | ED666: h+ leu1-32 ura4-D18 ade6-M210 | |
| strain, strain background (S. pombe) | rps23Δ | Bioneer | h+ ade6A14:E25-M210 or ade6-M216 ura4-D18 leu1-32 rps23Δ::kanMX4 | |
| strain, strain background (S. pombe) | rpl23Δ | Bioneer | h+ ade6-M210 or ade6-M216 ura4-D18 leu1-32 rpl23Δ::kanMX4 | |
| strain, strain background (S. pombe) | sre1Δ | Bioneer | h+ ade6-M210 or ade6-M216 ura4-D18 leu1-32 sre1Δ::kanMX4 | |
| strain, strain background (S. pombe) | rps25Δ | Bioneer | h+ ade6-M210 or ade6-M216 ura4-D18 leu1-32 rps25Δ::kanMX4 | |
| strain, strain background (S. pombe) | GST-rps2302-kanr | this study | PEY1816: h+ his3-D1 leu1-32 ura4-D18 ade6-M210 GST-rps2302- TADH1-PURA4-kanr-TTEF | |
| strain, strain background (S. pombe) | rps2302+-kanr | this study | PEY1817: h+ his3-D1 leu1-32 ura4-D18 ade6-M210 rps2302+- TADH1-PURA4-kanr-TTEF | |
| strain, strain background (S. pombe) | ofd1Δ sre1N | PMID: 18418381 | PEY873: h- his3-D1 leu1-32 ura4-D18 ade6-M210 ofd1Δ:: kanMX6 sre1(aa1-440) | |
| strain, strain background (S. pombe) | sre1N | PMID: 18418381 | PEY875: h- his3-D1 leu1-32 ura4-D18 ade6-M210 sre1(aa1-440) | |
| strain, strain background (S. pombe) | 2XSRE-ura4+ sre1N | PMID: 15797383 | PEY1290: h+ his-D1, leu1-32, ade6-M210, ura4-D18::2xSRE- ura4+-kanMX, sre1(aa1-440) | |
| strain, strain background (S. pombe) | 2XSRE-ura4+ sre1Δ | this study | PEY1489: h+ his-D1, leu1-32, ade6-M210, ura4-D18::2xSRE- ura4+-kanMX, sre1Δ::kanMX6 | |
| strain, strain background (S. pombe) | nro1Δ sre1N | PMID: 15797383 | PEY1410: h- his3-D1 leu1-32 ura4-D18 ade6-M210 nro1Δ:: kanMX4 sre1(aa1-440) | |
| strain, strain background (S. cerevisiae) | Yeast two-hybrid | PMID: 8978031 | AH109: MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MELTATA-lacZ | |
| recombinant DNA reagent | pSLF101 (vector) | PMID: 8332516 | ||
| recombinant DNA reagent | pART1 (vector) | PMID: 3034608 | ||
| recombinant DNA reagent | pGEX-4T3 (vector) | GE Healthcare Life Sciences | ||
| recombinant DNA reagent | pSLF172 (vector) | PMID: 9218719 | ||
| recombinant DNA reagent | pProEX-HTb (vector) | Invitrogen | ||
| recombinant DNA reagent | ppMAL-c5X (vector) | NEB | ||
| recombinant DNA reagent | EV bait (vector) | Clontech Matchmaker Gal4 Two-hybrid | Gal4_BD | |
| recombinant DNA reagent | EV prey (vector) | Clontech Matchmaker Gal4 Two-hybrid | Gal4_AD | |
| recombinant DNA reagent | human RPS23 (cDNA) | Duke Screening Center | ||
| recombinant DNA reagent | human OGFOD1 (cDNA) | Invitrogen Ultimate ORF | Invitrogen: 1OH23210 | |
| recombinant DNA reagent | nro1+ (plasmid) | PMID: 21481773 | Progenitors: PCR; pSLF101 vector | |
| recombinant DNA reagent | nro1 P6D, G8D, A11D, L15D (plasmids) | this study | Progenitors: nro1+ plasmid | |
| recombinant DNA reagent | rps2302+, P4D, G6D, A9D, L13D (plasmids) | this study | Progenitors: PCR; pART1 vector | |
| recombinant DNA reagent | GST-Rps23/Nro1 bacterial expression plasmids | this study | Progenitors: PCR; pGEX-4T3 vector | |
| recombinant DNA reagent | GST-rps2302 (plasmid) | this study | Progenitors: PCR; pSLF172 vector | |
| recombinant DNA reagent | GST (plasmid) | this study | Progenitors: PCR; pSLF172 vector | |
| recombinant DNA reagent | sre1N (plasmid) | PMID: 15797383 | Progenitors: PCR; pSLF101 vector | |
| recombinant DNA reagent | ofd1 bait (plasmid) | this study | Progenitors: PCR; Gal4_BD vector | |
| recombinant DNA reagent | OGFOD1 bait (plasmid) | this study | Progenitors: OGFOD1 cDNA; Gal4_BD vector | |
| recombinant DNA reagent | rps23 pombe prey (plasmid) | this study | Progenitors: PCR; Gal4_AD vector | |
| recombinant DNA reagent | RPS23 human prey (plasmid) | this study | Progenitors:RPS23 cDNA; Gal4_AD vector | |
| recombinant DNA reagent | MBP-Rps23 (plasmid) | this study | Progenitors: PCR; pMAL-c5X vector | |
| recombinant DNA reagent | ofd1 bacterial expression plasmid | PMID: 21481773 | Progenitors: PCR; pProEX-HTb vector | |
| recombinant DNA reagent | nro1 bacterial expression plasmid | PMID: 21481773 | Progenitors: PCR; pProEX-HTb vector | |
| recombinant DNA reagent | 6xHis-RPS23 (plasmid) | this study | Progenitors: RPS23 cDNA; pProEX-HTb vector | |
| antibody | IRDye800CW/IRDy e680RD secondaries | LI-COR | (1:20000) | |
| antibody | anti-Nro1 (rabbit polyclonal) | PMID: 15797383 | (1:10000) | |
| antibody | anti-Dsc5 (rabbit polyclonal) | PMID: 22086920 | (1:10000) | |
| antibody | anti-Sre1 (rabbit polyclonal) | PMID: 15797383 | (1:2500) | |
| antibody | anti-Ofd1 (rabbit polyclonal) | PMID: 18418381 | (1:10000) | |
| antibody | anti-GST (mouse monoclonal) | Covance | Covance:MMS-112R | (1:1000) |
| antibody | anti-GFP (mouse monoclonal) | Roche Applied Science | Roche:1814460 | (1:1000) |
| antibody | anti-Rps23 (mouse monoclonal) | Santa Cruz | Santa Cruz:sc-100837 | (1:200) |
| antibody | anti-Rps5 (mouse monoclonal) | Santa Cruz | Santa Cruz:sc-390935 | (1:100) |
| antibody | anti-actin (mouse monoclonal) | Santa Cruz | Santa Cruz:sc-47778 | (1:200) |
| Chemical compound, drug | DMOG | Frontier Scientific | Frontier Scientific:D1070 | |
| Chemical compound, drug | DSP | Thermo Fisher Pierce | Thermo Fisher Pierce:22586 | |
| Chemical compound, drug | Heavy lysine | Cambridge Isotope Laboratories | Cambridge Isotope Laboratories: CNLM-291-H-1 | |
| Chemical compound, drug | LMB | Cell Signaling | Cell Signaling:9676 |
Additional files
-
Supplementary file 1
Yeast strains
Genotypes and references for the yeast strains used in the study.
- https://doi.org/10.7554/eLife.28563.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28563.017





