Assessing the danger of self-sustained HIV epidemics in heterosexuals by population based phylogenetic cluster analysis
Figures
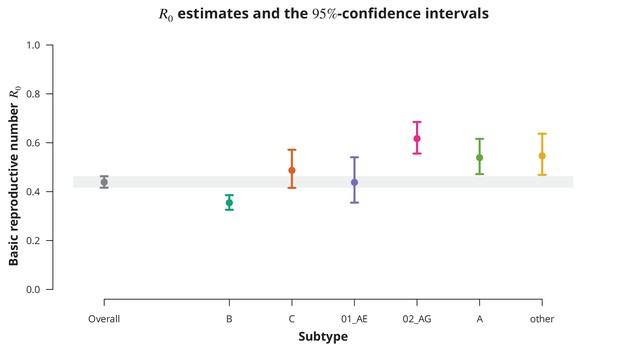
Overall basic reproductive number and per subtype from stratified analysis.
The dark gray point indicates the overall basic reproductive number estimate (by neglecting the transmission chain subtypes) and the corresponding -confidence interval is shown with the dark gray line and the gray-shaded band. The analogous results from the per-subtype stratified analysis are represented by colored points and lines, each color corresponding to one of the subtypes (B, C, CRF01_AE, CRF02_AG or A) or the group of subtypes (other).
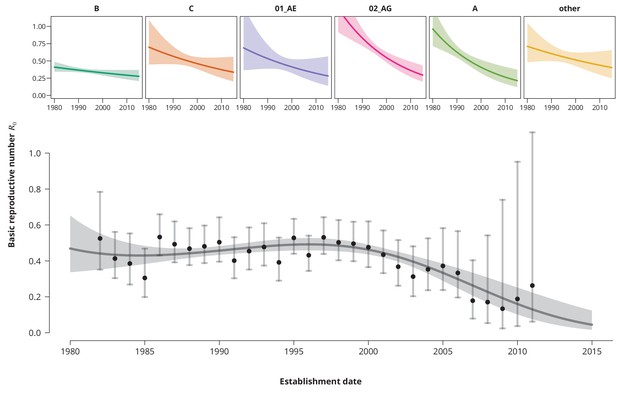
Time trends for .
The upper smaller panels show the time trends for from the subtype-stratified analyses, in which the ’s were modeled as linear functions of establishment date (i.e., for each subtype the time trend rate was assumed to be constant). The colored shaded-bands correspond to the -prediction bands. The (best-fitting) nonlinear time trend for from the overall analysis is displayed in the lower panel (dark gray curve) together with the -prediction band (gray-shaded area). The black points represent the estimates from the per establishment year stratified analyses and the gray vertical lines the corresponding -confidence intervals.
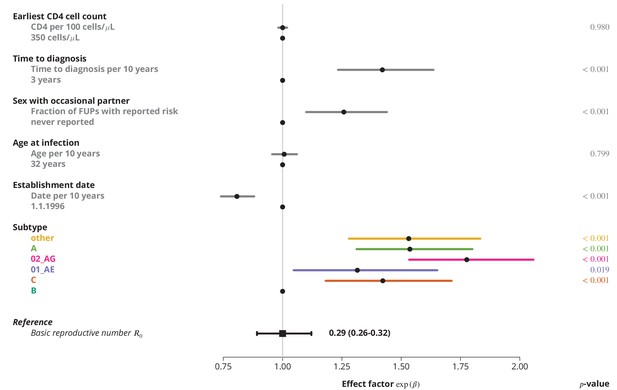
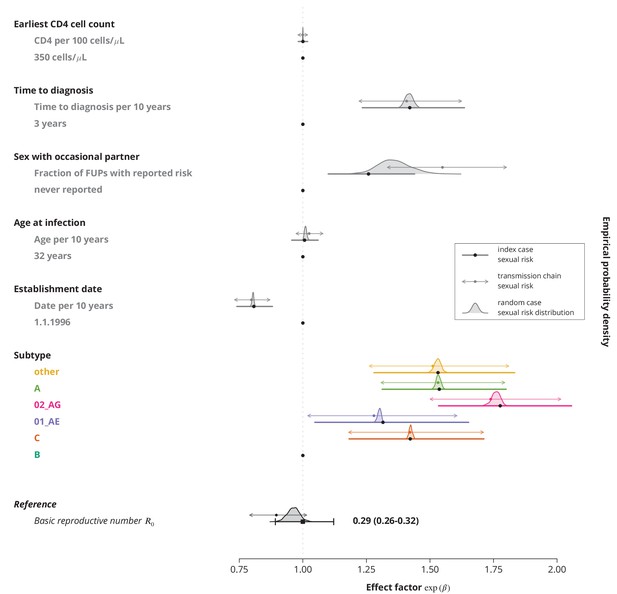
Effect of different factors on the basic reproductive number from the multivariate model with only linear factor terms.
The black square and the black line show the reference basic reproductive number and its -confidence interval (for a transmission chain of subtype B which started on 1.1.1996, and in which the index case was diagnosed 3 years after the infection, was 32 years old upon infection, never reported on having sex with occasional partner and had the earliest CD4 cell count of 350 cells per μL). The vertical gray line separates the factors associated with lower (left; effect factor ) and from the factors contributing to higher (right; effect factor ). The black points on this line refer to the reference transmission chain. The colored and dark gray lines represent the effect sizes from multivariate model (black circles depicting the estimates) for different factors and their -confidence intervals. The corresponding -values are shown in the rightmost column. FUP, follow-up visit.
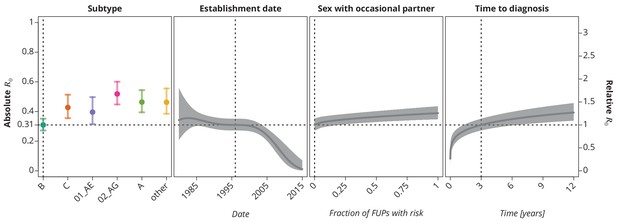
Final multivariate model’s profile plots of factors associated with the basic reproductive number .
The vertical dotted lines depict the reference transmission chain (of subtype B, started on 1.1.1996, in which the observed index case did not report having sex with occasional partner and was diagnosed after 3 years after the infection). The left -axis represents the basic reproductive number whereas the right -axis corresponds to the relative values of as compared to the baseline . The as the function of specific factor (with the other factors held fixed at the reference value) is displayed by the colored (for HIV-1 subtype) and the dark gray (establishment date, sexual risk behavior and time to diagnosis) lines. The vertical bars and the shaded bands, respectively, correspond to the -confidence intervals.
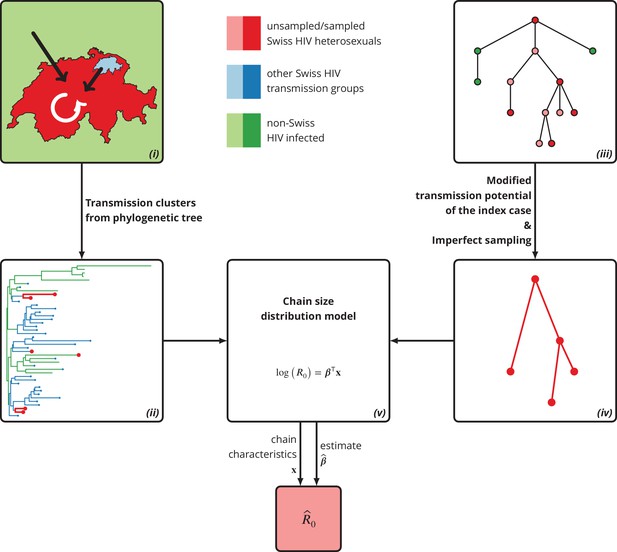
Graphical representation of our phylogeny-based statistical approach.
(i): HIV transmission among heterosexuals in Switzerland (white arrow) has never led to a self-sustained epidemic. However, the unknown potential of imported infections (black arrows) either from abroad or from other transmission groups in Switzerland remains a large concern. (ii): The HIV transmission chains corresponding to Swiss heterosexuals (depicted in red) were identified from the phylogenetic tree containing the SHCS and background viral sequences. (iii): Our mathematical model is based on the discrete-time branching process with nodes of three different types: sampled Swiss infection (red), unsampled Swiss infection (light red) and foreign infection infected by a Swiss index case before moving to Switzerland (green). (iv): Our method for inferring accounts for both imperfect sampling and modified transmission potential of the index case. (v): Moreover, it includes the baseline transmission chain characteristics to assess the determinants of .
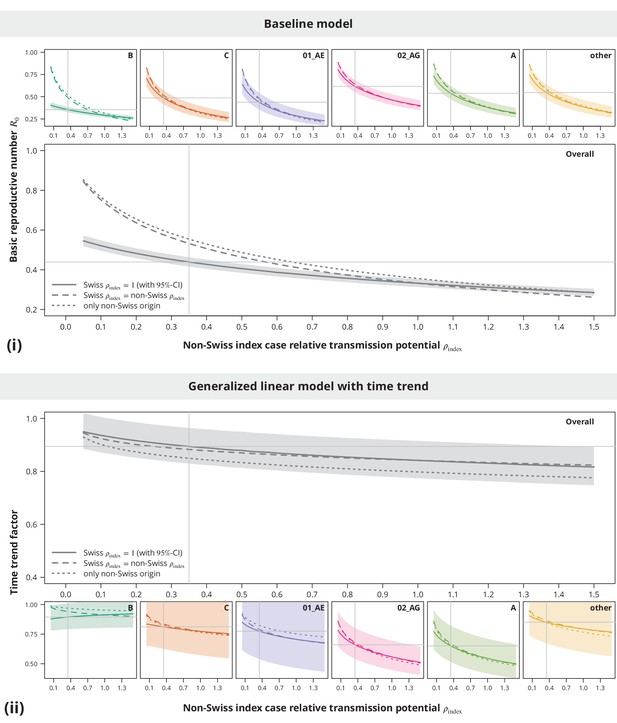
Sensitivity analysis regarding the index case relative transmission potential.
Panel (i) shows the sensitivity of the estimates from baseline model and panel (ii) the sensitivity of the time trend factor. The colored lines represent the subtype-stratified analyses, while the results from the overall models are shown in gray. In the first sensitivity analysis, the of Swiss-originating transmission chains was held at and the of non-Swiss origin varied (solid lines). In the second analysis, the of Swiss and non-Swiss origin was the same (dashed lines). The dotted lines show the results from the sensitivity subanalysis including only the transmission chains of non-Swiss origin. The vertical and horizontal lines depict the parameters and estimates from the main analysis, respectively.
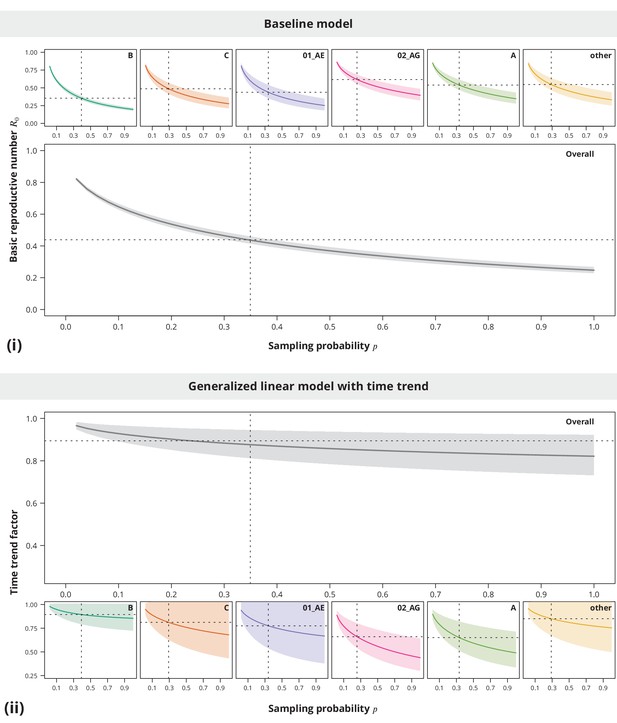
Sensitivity analysis regarding the sampling density.
The index case relative transmission potential parameter was the same as used in the main analyses, while the sampling densities varied (-axis). In the pooled analysis (larger plots) the sampling density was the same for all transmission chains. Panel (i) shows the corresponding estimates of the basic reproductive number and the time trend factor estimates are displayed in panel (ii). The dotted vertical lines depict the sampling densities used for each subtype in our study (subtype-stratified plots) and the mean sampling density over all transmission chains (overall plots). The horizontal dotted lines represent the estimates from the main analysis.
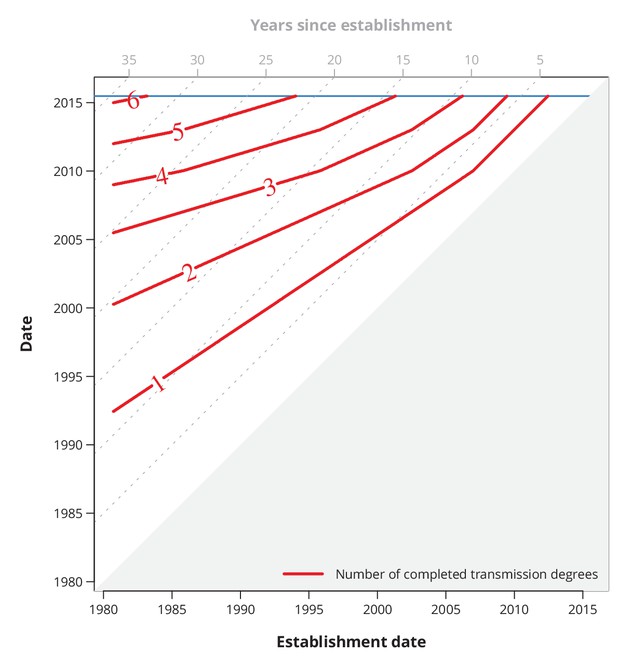
Conservative (with respect to ongoing transmission) maximum number of completed transmission degrees by a given date.
The red lines show the date (-axis) by which at least a certain number (red numbers) of transmission degrees have been completed for a transmission chain with a specific establishment date (-axis). The diagonal dotted gray lines depict the number of years since the establishment date, and the horizontal blue line represents the last sampling date.
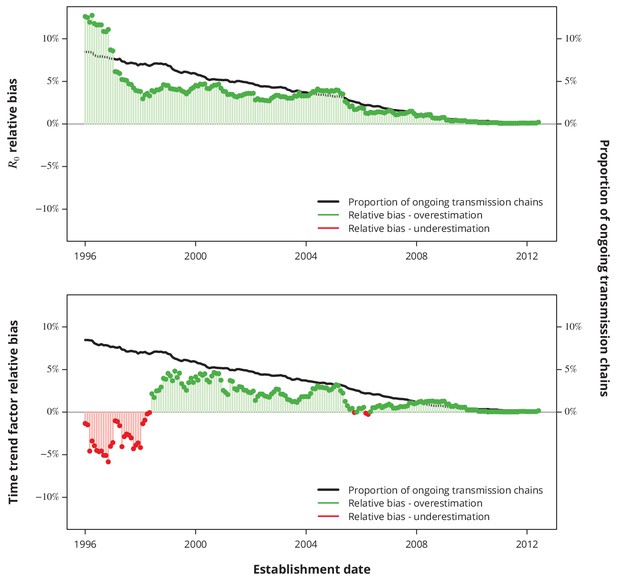
Relative bias due to ongoing transmission.
The upper panel shows the relative bias of the basic reproductive number from the baseline model and the lower panel the relative bias of the linear time trend factor from the corresponding generalized linear model. The proportion of active transmission chains over time is represented by the black line. The relative bias associated with overestimation and underestimation is displayed with green and red bars-points, respectively. Absence of bias is depicted by the horizontal gray lines.
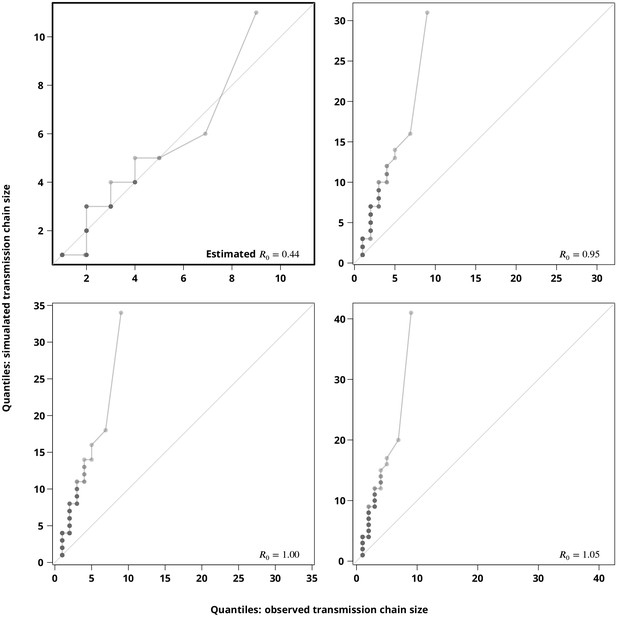
Sensitivity analysis regarding the stuttering transmission chains assumption.
The Q-Q plots compare the hypothetical transmission chain size distributions (-axis showing their empirical permilles) with the transmission chain size distribution (empirical permilles on the -axis) inferred from the phylogeny. The upper left plot compares the distribution of the simulated transmission chain sizes based on the estimated with the (from the phylogeny) observed transmission chain sizes and thus verifies the estimate. The remaining plots compare the simulated transmission chain size distributions against the extracted transmission chain sizes for closer to to justify the subcritical transmission assumption. Each point represents a permille, hence the darker points indicate more overlapping permilles.
Comparison of effect sizes in the multivariate model with linear terms only for different sexual risk behavior definitions of a transmission chain.
The thick lines with black circles show the original effect sizes (where the index case determined the sexual risk behavior of the transmission chain) and their -confidence intervals. The empirical distribution of the effect sizes where a random individual in a transmission chain determines its sexual risk behavior is displayed by the shaded areas. The thinner horizontal double sided arrows with the filled circles correspond to the effect sizes and their -confidence intervals for the transmission chain level fraction of follow-up visits (FUPs) with reported sex with occasional partner by any of the infected individuals from the transmission chain. The vertical dotted gray line depicts the reference from the original model, i.e., using the index case to define the sexual risk behavior.
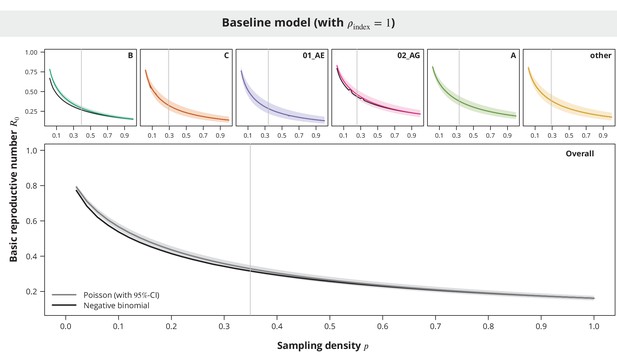
Comparison between the Poisson and the negative binomial offspring distribution baseline model estimates.
The dark gray and colored lines show the estimates from the model with Poisson offspring distribution, while the black lines correspond to the negative binomial distribution. The index case relative transmission potential parameter was fixed to and the sampling density (-axis) varied. In the overall analysis the sampling density was the same for all transmission chains regardless of their subtype. The vertical gray lines depict the sampling densities used for each subtype in our study (above panels) and the mean sampling density in the overall analysis (bottom panel).
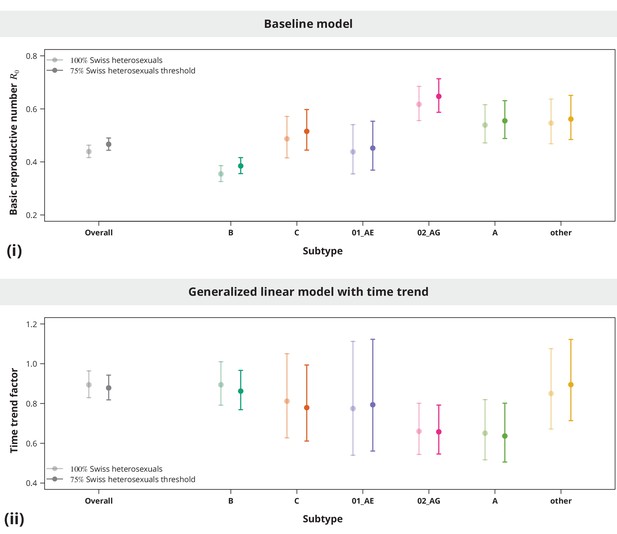
Sensitivity analysis regarding the transmission cluster definition.
The upper panel (i) compares the estimated with the original cluster definition (brighter lines) with the estimated based on the relaxed cluster definition (darker lines) from the overall analysis (in gray) and subtype-stratified analyses (in colors). Similarly, the bottom panel (ii) shows the comparison between the estimated time trend factors obtained from the transmission chain sizes based on different cluster definition thresholds.
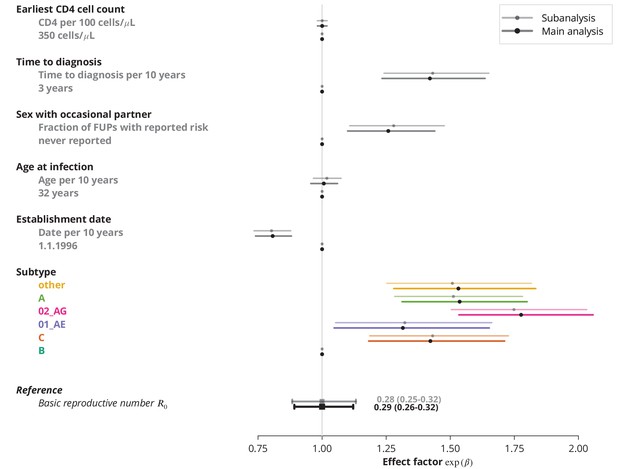
Subanalysis for the transmission chains with available follow-up information about sex with occasional partner of the index case compared to the main analysis with imputed data.
The effect sizes from the subanalysis are shown in brighter colors and those from the main analysis in dark. In the main analysis, the missing data were replaced by never reporting sex with an occasional partner.
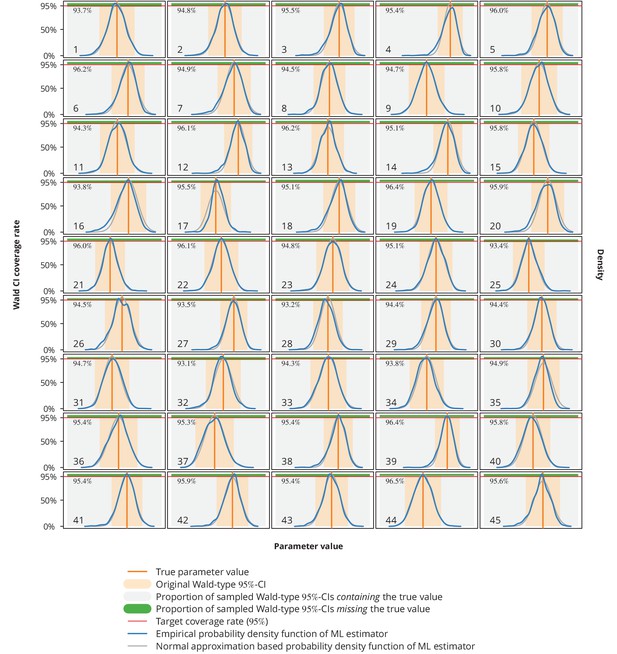
Empirical distribution of maximum likelihood (ML) estimator and the Wald-type confidence intervals (CI) coverage rates.
Each plot represents a single parameter from a single model (see Appendix 1—table 1 for the parameters overview including their values), where the number in the lower left corner denotes the parameter’s consecutive parameter number. The light gray-shaded area represents the proportion of the Wald-type -CIs from the parametric bootstrap simulations which contained the true value (depicted by the vertical orange line), while the green-shaded area corresponds to those CIs from the simulations that missed the true value. The numbers in the upper left corners are the coverage rates from the parametric bootstrap. The original Wald -CIs used in our study are displayed with the light orange-area. The dark blue and gray lines show the empirical distribution of ML estimators from the parametric bootstrap samples and the normal approximation based probability density function, respectively. The horizontal red lines depict the target coverage rate of .
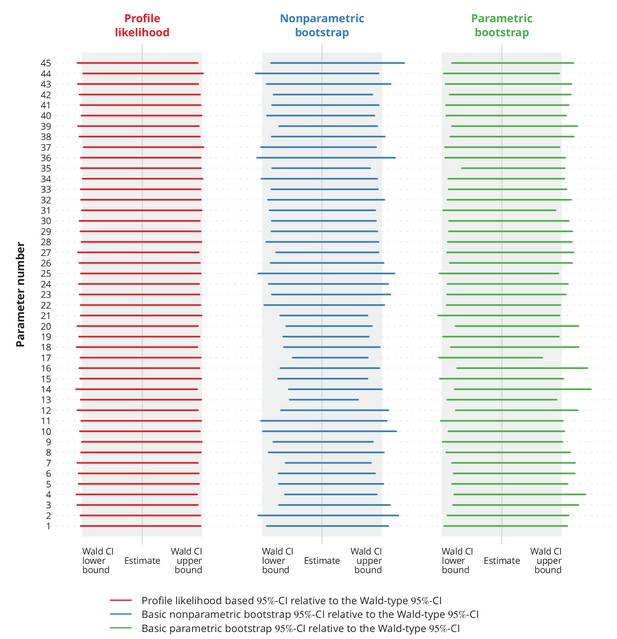
Comparison of different types of -confidence intervals (CI) with the normal approximation based Wald-type -CIs.
Each column corresponds to a different type of CIs, namely the profile likelihood based CIs, the basic nonparametric bootstrap CIs and the basic parametric bootstrap CIs. Each row represents a single parameter (the overview of the parameters is provided in Appendix 1—table 1). The colorful lines show the specific CIs compared to the corresponding Wald-type CIs, namely their relative widths and positions. The gray-shaded areas represent the Wald-type -CIs.
Tables
Transmission chain size distribution and model parameters.
https://doi.org/10.7554/eLife.28721.003| Subtype | Overall | ||||||
|---|---|---|---|---|---|---|---|
| B | C | 01_AE | 02_AG | A | Other | ||
| Total number of chains, (%) | 1643 (53%) | 322 (10%) | 239 (7.7%) | 331 (11%) | 327 (11%) | 238 (7.7%) | 3100 (100%) |
| Chain size, (%) | |||||||
| 1 | 1437 (87%) | 280 (87%) | 206 (86%) | 272 (82%) | 269 (82%) | 195 (82%) | 2659 (86%) |
| 2 | 158 (9.6%) | 34 (11%) | 31 (13%) | 40 (12%) | 44 (13%) | 36 (15%) | 343 (11%) |
| 3 | 30 (1.8%) | 7 (2.2%) | 1 (0.42%) | 10 (3.0%) | 10 (3.1%) | 6 (2.5%) | 64 (2.1%) |
| 4 | 12 (0.73%) | - | 1 (0.42%) | 6 (1.8%) | 3 (0.92%) | 1 (0.42%) | 23 (0.74%) |
| 5 | 1 (0.06%) | 1 (0.31%) | - | 2 (0.6%) | 1 (0.31%) | - | 5 (0.16%) |
| 6 | 1 (0.06%) | - | - | 1 (0.3%) | - | - | 2 (0.06%) |
| 7 | 1 (0.06%) | - | - | - | - | - | 1 (0.03%) |
| 8 | 2 (0.12%) | - | - | - | - | - | 2 (0.06%) |
| 9 | 1 (0.06%) | - | - | - | - | - | 1 (0.03%) |
| Sampling probability, (SD) | 0.39 | 0.29 | 0.34 | 0.26 | 0.33 | 0.29 | 0.35 (0.05) |
| Chain origin, (%) | |||||||
| Swiss () | 948 (58%) | 36 (11%) | 36 (15%) | 36 (11%) | 47 (14%) | 30 (13%) | 1133 (37%) |
| non-Swiss () | 695 (42%) | 286 (89%) | 203 (85%) | 295 (89%) | 280 (86%) | 208 (87%) | 1967 (63%) |
Patients’ demographic characteristics.
https://doi.org/10.7554/eLife.28721.006| Patients | Transmission chains | |
|---|---|---|
| Index case | ||
| Total number, | 3698 | 3100 |
| Age at estimated date of infection [in years], median (IQR) | 29.2 (23.1—37.8) | 28.8 (22.8—37.4) |
| Estimated date of infection, median (IQR) | Jun 1996 (Sep 1990—Nov 2001) | Nov 1995 (Sep 1989—May 2001) |
| Time to diagnosis [in years], median (IQR) | 3.40 (1.66—5.24) | 3.54 (1.78—5.43) |
| Reported sex with occasional partner [as fraction of FUPs*], median (IQR) | 0.53 (0.09—0.89) | 0.50 (0.07—0.88) |
| No available FUP†, (%) | 250 (6.8%) | 226 (7.3%) |
| Earliest CD4 count [per μL]‡, median (IQR) | 310 (143—510) | 300 (134—507) |
-
*Follow-up visit (FUP).
†Patients without FUP questionnaire regarding the sexual risk behavior. See Sensitivity analyses.
-
‡One patient did not have any available CD4 cell count. The missing value was imputed with the mean CD4 cell count.
Overview of all the parameters, their estimates and the -confidence intervals fitted in all the models presented in this study.
https://doi.org/10.7554/eLife.28721.021| Subtypes | Parameter number | Parameter name | Parameter estimate | Wald-type -CI | Profile likelihood -CI |
|---|---|---|---|---|---|
| Overall | 1 | ||||
| B | 2 | ||||
| C | 3 | ||||
| 01_AE | 4 | ||||
| 02_AG | 5 | ||||
| A | 6 | ||||
| other | 7 | ||||
| Overall | 8 | ||||
| 9 | |||||
| B | 10 | ||||
| 11 | |||||
| C | 12 | ||||
| 13 | |||||
| 01_AE | 14 | ||||
| 15 | |||||
| 02_AG | 16 | ||||
| 17 | |||||
| A | 18 | ||||
| 19 | |||||
| other | 20 | ||||
| 21 | |||||
| Overall | 22 | ||||
| 23 | |||||
| 24 | |||||
| Overall | 25 | ||||
| 26 | |||||
| 27 | |||||
| 28 | |||||
| 29 | |||||
| 30 | |||||
| 31 | |||||
| 32 | |||||
| 33 | |||||
| 34 | |||||
| 35 | |||||
| Overall | 36 | ||||
| 37 | |||||
| 38 | |||||
| 39 | |||||
| 40 | |||||
| 41 | |||||
| 42 | |||||
| 43 | |||||
| 44 | |||||
| 45 |
Establishment date models obtained with the AIC/BIC forward selection and backward elimination and their respective AIC and BIC values as well as the -values from the likelihood ratio test compared to the null model without any covariates.
Terms that were part of the respective final model are marked by .
| AIC | BIC | |||
|---|---|---|---|---|
| Forward | Backward | Forward | Backward | |
| AIC | ||||
| BIC | ||||
| -value from LR test | ||||
Multivariate models obtained with the AIC/BIC forward selection and backward elimination algorithms.
The terms listed in the table are the terms identified from the single determinant model selections and the crosses indicate the terms entering the multivariate models. The null model from the likelihood ratio test refers to the baseline model without any covariates (not even the subtype).
| AIC | BIC | |||
|---|---|---|---|---|
| Forward | Backward | Forward | Backward | |
| AIC | ||||
| BIC | ||||
| -value from LR test | ||||
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28721.010





