LARP4 mRNA codon-tRNA match contributes to LARP4 activity for ribosomal protein mRNA poly(A) tail length protection
Figures
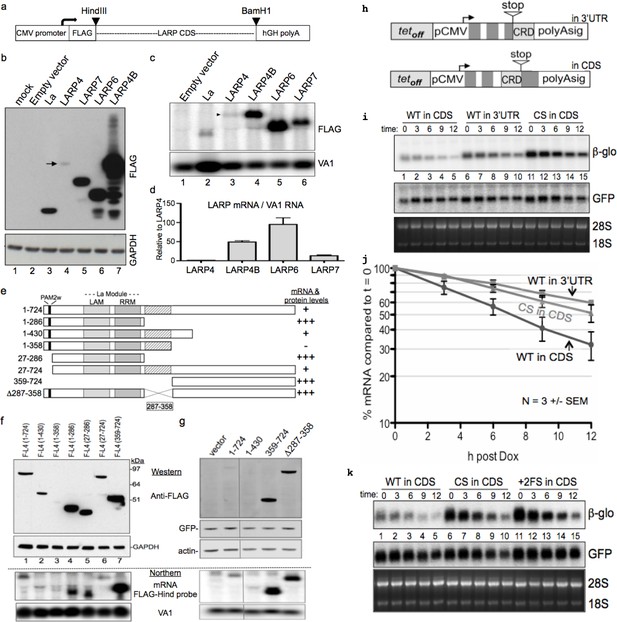
LARP4 mRNA contains a codon-specific, coding region determinant (CRD) of instability in that limits expression.
(a) Schematic showing expression cassette in pFLAG-CMV2 plasmids that differ only in the open reading frame (ORF) coding sequences (CDS). (b) Western blot using anti-FLAG antibody; arrow indicates LARP4 band, lane 4. (c) Northern blot, upper panel shows detection by FLAG-Hind antisense probe. VA1 RNA was for transfection control and quantification. (d) Normalized expression of three experiments (n = 3), error bars represent s.e.m.; Flag-LARP4 set to 1. (e) Schematic of LARP4 mutated constructs transfected with VA1 for f and g. (f) Upper: Western blot with anti-FLAG and anti-GAPDH. Lower: Northern blot with FLAG-Hind antisense and VA1 probes. (g) Upper: Western blot with anti -FLAG, -actin and -GFP antibodies. Lower: Northern for FLAG-Hind and VA1. (h) Schematic showing two β-globin reporters containing CRD constructs (see text); 'in 3’UTR' following the stop codon, and in frame preceding the stop 'in CDS'. (i) Northern blot time course of decay of the β-glo CRD reporter mRNAs. WT = wild type CRD sequence and CS = synonymous codon swapped version of the CRD. (j) Quantification of β-glo mRNA in i; 3 independent experiments, error bars represent s.e.m. GFP used for normalization, and each t = 0 was set to 100%. (k) Similar to i; +2 FS= + 2 frameshift version of the WT CRD.
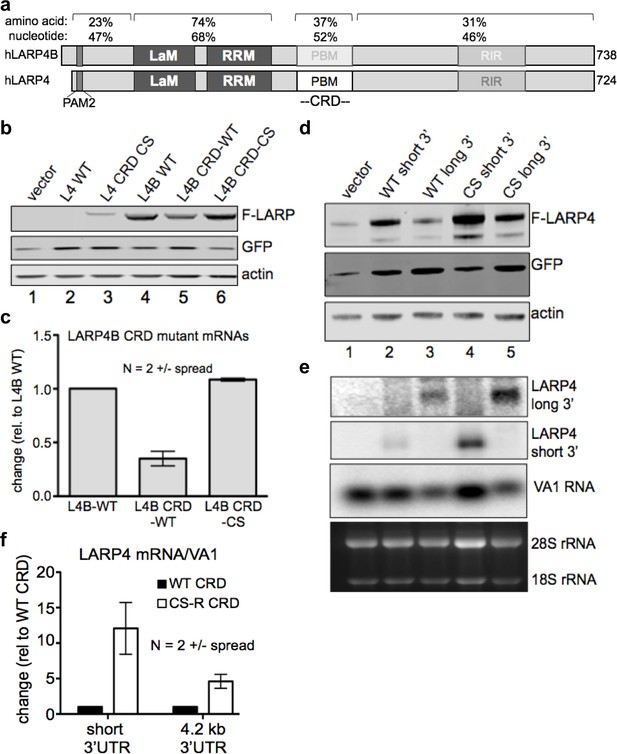
The LARP4 CRD confers instability when transferred to a mRNA of high expression level and in the context of its natural regulatory 3' UTR.
(a) Diagram of human LARP4 and LARP4B with nucleotide and amino acid sequence identities of relevant subregions: PAM2 = PAM2, LaM = La Motif, RRM = RNA Recognition Motif, PBM = PABP interaction motif. (b–c) LARP4 CRD replaced the homologous region of LARP4B as either WT or the CS-B form (see text). Western blot b, and northern blot of F-LARP4B mRNA c, the latter quantified after two independent experiments; WT was set to 1 and normalization was to VA1, error bars = spread. (d-e) The 4.2 kb 3’UTR of LARP4 was inserted at the 3’ end of the LARP4 WT or CS-R constructs (see below). Western blot d, and northern blot e. (f) Quantification of northern data from two independent experiments; error bars = the spread.
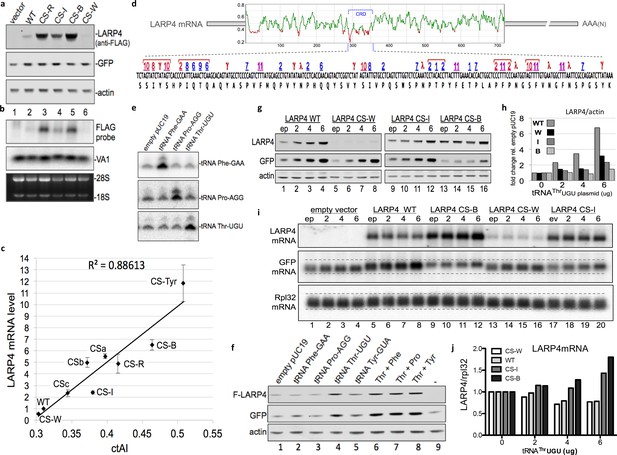
CRD synonymous codon match to limiting cellular tRNA levels and wobble dynamics control LARP4 expression levels.
(a) Western blot of proteins from LARP4 codon swap (CS) constructs. WT = wild type LARP4; others are described in the text. (b) Northern blot of RNA from same cells in a. (c) Cellular tRNA index (ctAI) scores of the CRD regions of CS constructs. ctAI scores plotted on x-axis and Flag-LARP4 mRNA levels relative to Flag-LARP4-WT on the y-axis; N = 7 biological replicates for CS-R and CSb, N = 5 biological replicates for CS-W, CS-I, and CS-B, N = 4 biological replicates for CSc, N = 3 biological replicates for CS-Tyr and CSa; error bars reflect the s.e.m. (d) Top: Sliding window ctAI-based score plot depiction of the full length LARP4 ORF of 724 codons (numbered on X-axis). Regions scoring below 0.4 (Y-axis) are colored red (see text). The position of the CRD codons 287–358 are indicated by the bracket. Bottom: The CRD codons are shown with single letter amino acids underneath and with numbers above that correspond to their cognate tRNAs as follows. Numbers 1–11 designate the rank order low level tRNAs in Bin-1 from Table 1; red = wobble decoded, blue = Watson:Crick decoded. Symbols γ and λ designate weak (all-A + T) codons that must be wobble decoded (see text) by non-Bin-1 tRNAs; note that #11 (F, Phe) is also a weak (all-A + T) codon. (e) Northern blot 48 hr after tRNA gene-containing plasmids were transfected into HEK293 cells. (f) Western blot of extracts from HEK293 cells transfected with empty pUC19 or plasmids containing tRNA genes for PheGAA, ProAGG, ThrUGU or combinations as indicated above the lanes, together with empty pCMV2 or F-LARP4-WT, and GFP. Antibodies used are indicated to the left of the panels. (g–j) HEK293 cells transfected with empty plasmid (ep) or 2, 4, or 6 ug of tRNAThrUGU plasmid together with empty pCMV2 (empty vector), F-LARP4-WT, CS-W, CS-I, or CS-B, and GFP. (g) Western blots as indicated. (h) Quantification of LARP4 in g as indicated. (i) Northern blot. (j) quantification of LARP4 mRNA/Rpl32 mRNA from northern blot in i.

Sequences of the CRD regions of the LARP4 CS constructs.
https://doi.org/10.7554/eLife.28889.006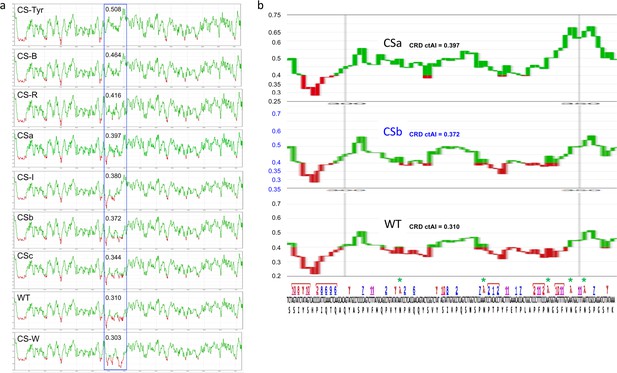
Plots of 10 nt sliding window ctAI translation proxy scores of the full length LARP4 constructs.
(a) The plot of each construct is shown with its CRD region in the blue rectangle with its ctAI score. Note that the Y-axis translation proxy score lines may be different in different plots. (b) The CRD regions of three plots were expanded and aligned underneath with the codon sequence of the WT CRD from Figure 3d. The green asterisks denote Asn codons that underwent AAU to AAC substitution from the CSb to the CSa construct. Again, note the difference in the Y-axes.
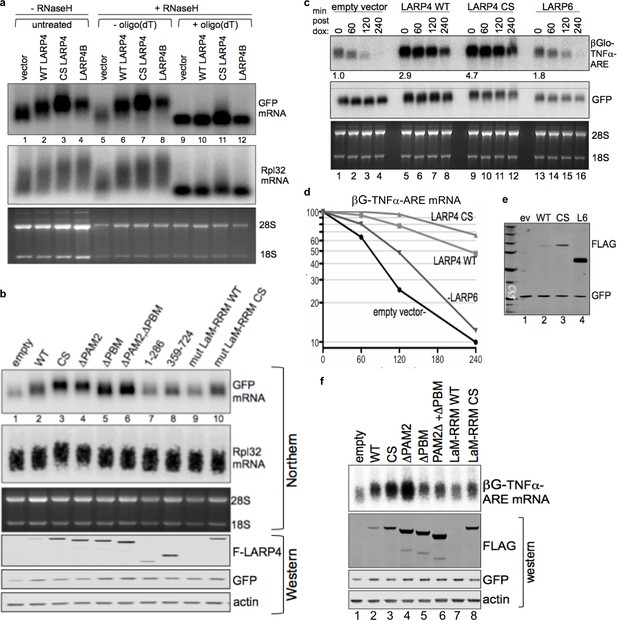
CRD-mediated increase in LARP4 leads to heterologous mRNA 3' PAT lengthening and stabilization dependent on its PABP- and RNA- interaction domains.
(a) Northern blot after RNase H ± oligo(dT) treatment of total RNA from HEK293 cells transfected with constructs indicated above the lanes; CS = CS R version of the CRD in full length LARP4. (b) Upper: northern blot for GFP mRNA mobility shift activity of LARP4 constructs some of which contain the CS-R version of the CRD. WT = wild type, CS = full length LARP4 with CS-R CRD, ΔPAM2 = PAM2 deleted w/CS R, ΔPBM = PBM/CRD deleted, ΔPAM2ΔPBM, mut LaM-RRM WT and mut LaM-RRM CS = previously described M3 point mutations in the LaM and RRM in the full length LARP4 WT and CS-R versions respectively. Lower: western blot. (c) Northern blot of mRNA decay time course of HeLa Tet-Off cells transfected with βG-TNFα-ARE, GFP and either empty vector, LARP4-WT, LARP4-CS-R or LARP6. Cells harvested after 0, 60, 120 and 240 mins after doxycycline. Numbers under lanes for t = 0 indicate quantification of βG mRNA divided by GFP mRNA in the same lane, with lane 1 set to 1.0. (d) Quantification of βG-TNFα-ARE mRNA from c; the t = 0 for each was set to 100%. (e) Western blot of proteins tested in c, d. (f) Northern blot of HEK293 cells after transfection with βG-TNFα-ARE (constitutive promoter), GFP and either empty pCMV2 or F-LARP4-WT or mutants indicated above lanes as described for b; three lower panels show western blot of extracts using antibodies as indicated.
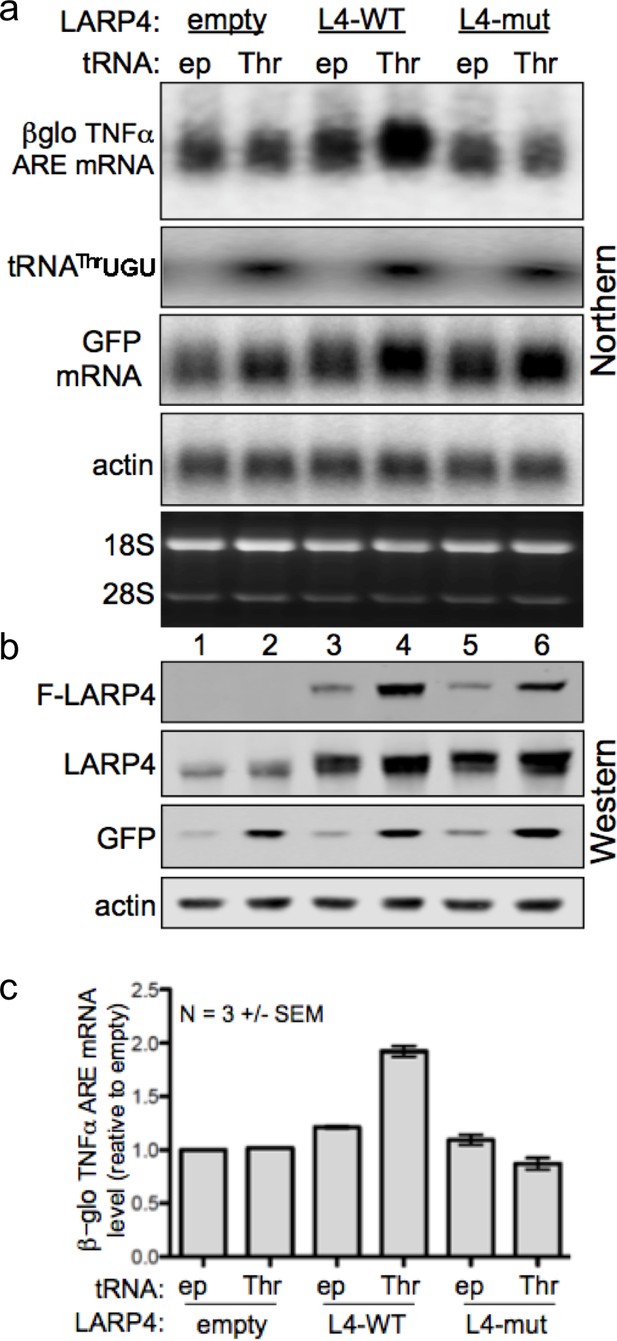
Increasing levels of the low abundance tRNAThrUGU elevates LARP4 with consequent PAT lengthening and stabilization of heterologous mRNAs.
(a) Northern blot of HEK293 cells after transfection with βG-TNFα-ARE (constitutive promoter), GFP and either empty pUC19 (ep) or tRNAThrUGU plasmid together with empty pCMV2 or F-LARP4 (L4–WT) or L4-mut M3 containing mutated LaM-RRM. Other panels show probings for tRNAThrUGU, GFP mRNA and actin mRNA. (b) Western blot of extracts from the cells in a; anti-FLAG for top panel, anti-LARP4 for second panel. (c) Quantification of βG-TNFα-ARE mRNA in a from 3 independent experiments; error bars = s.e.m.
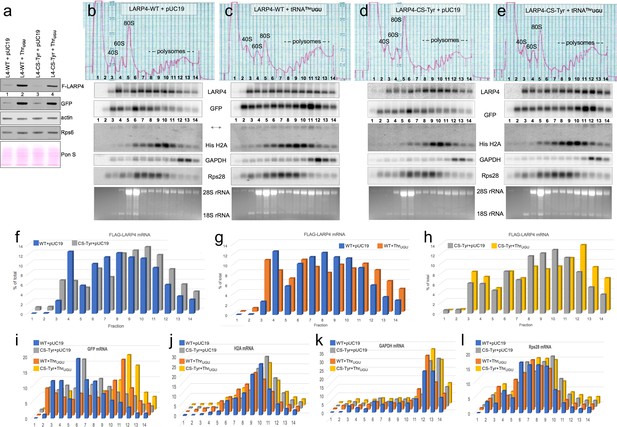
LARP4. Synonymous codon substitutions to the CRD and tRNAThrUGU promote apparent translation efficiency of LARP4 as monitored by polysome sedimentation analysis.
(a) Western blot of HEK293 cells after transfection with GFP and either empty pUC19 or tRNAThrUGU plasmid together with F-LARP4 (L4–WT) or F-LARP4 CS-Tyr (L4-CS-Tyr) The total amount of plasmid transfected in each was the same in all four samples. (b–e) Polysome sedimentation profiles of the same extracts as in a. RNAs from each of the numbered fractions was analyzed by northern blot using the probes indicated next to the panels. The lower panels show ethidium bromide staining of the gels. (f–h) quantitations of FLAG-LARP4 from the northern blots in b-e. (i–l) Quantitations of different mRNAs in b-e as indicated above the graphs.
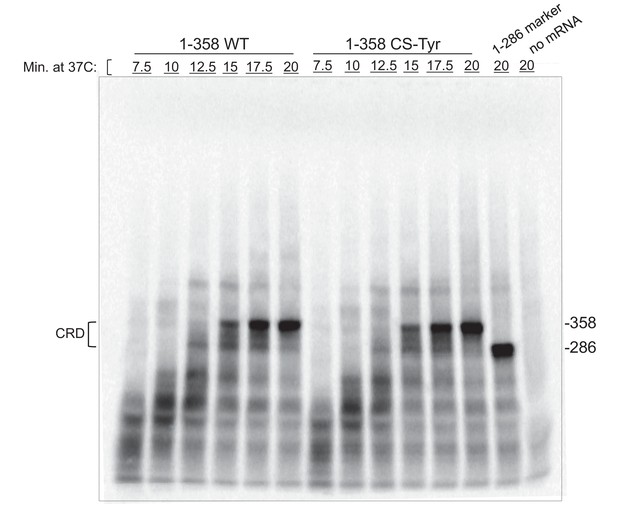
Time course in vitro translation of T7-synthesized mRNAs.
−358 and −286 at right indicate positions of N- and C- terminal boundaries of the CRD, as indicated by bracket on the left. The 358 size bands resulted from in vitro translation of the T7-synthesized mRNAs (see Materials and methods below and text).
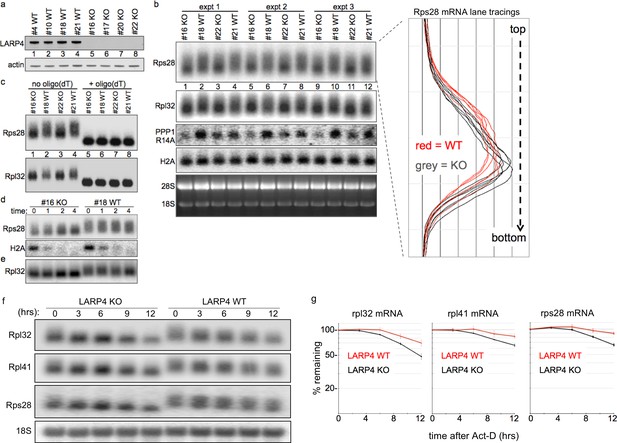
LARP4 gene-deleted knockout (KO) cells exhibit decreased 3' PAT length and stability of ribosome protein mRNAs.
(a) Western blot of LARP4 from independent isolates of WT and LARP4 KO MEFs. (b) Northern blot from 4 MEF cell lines in a; probes as indicated to the left of the panels, Rps28, Rpl32, PPP1R14A, histone H2A mRNAs, and EtBr stained gel. Densitometric lane tracings for each lane of a Rps28 exposure is shown to the right as indicated. (c) RNase H assay in presence or absence of oligo(dT) as indicated. (d e,) Time course of mRNA decay in LARP4 KO and WT MEFs after transcription shut-off (in hrs) by actinomycin-D, probed for Rps28, Rpl32 and histone H2A mRNAs as indicated to the left; e contains the same RNA preparation as in d but run on a separate gel. (f) Northern blot of 12 hr act-D time courses for LARP4 KO and WT MEFs probed for the RNAs indicated to the left. (g) Graphs showing quantifications of duplicate experiments including panels in f, as indicated. The mRNA quantification at each time was normalized against 18S rRNA in the same lane. Error bars at each time point reflect the spread of the duplicates.
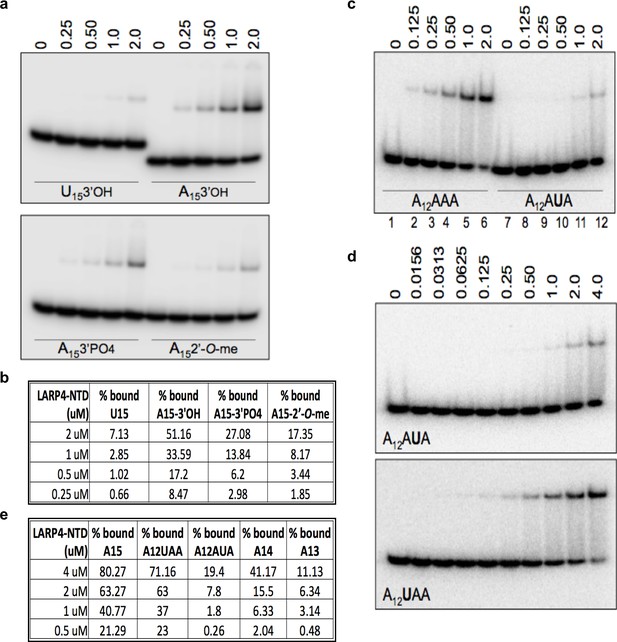
The N-terminal LaM-RRM module-containing fragment of LARP4 exhibits poly(A) 3' end sensitivity.
(a, c and d) Electrophoretic mobility shift assay (EMSA) using purified recombinant LARP4-NTD (1-286) protein in varying concentrations indicated above the lanes in uM with the purified oligo-RNA species indicated in the lower part of the gels. (b and e) Quantification of binding in a, c and d, and EMSA experiments with other oligo-RNAs as indicated.
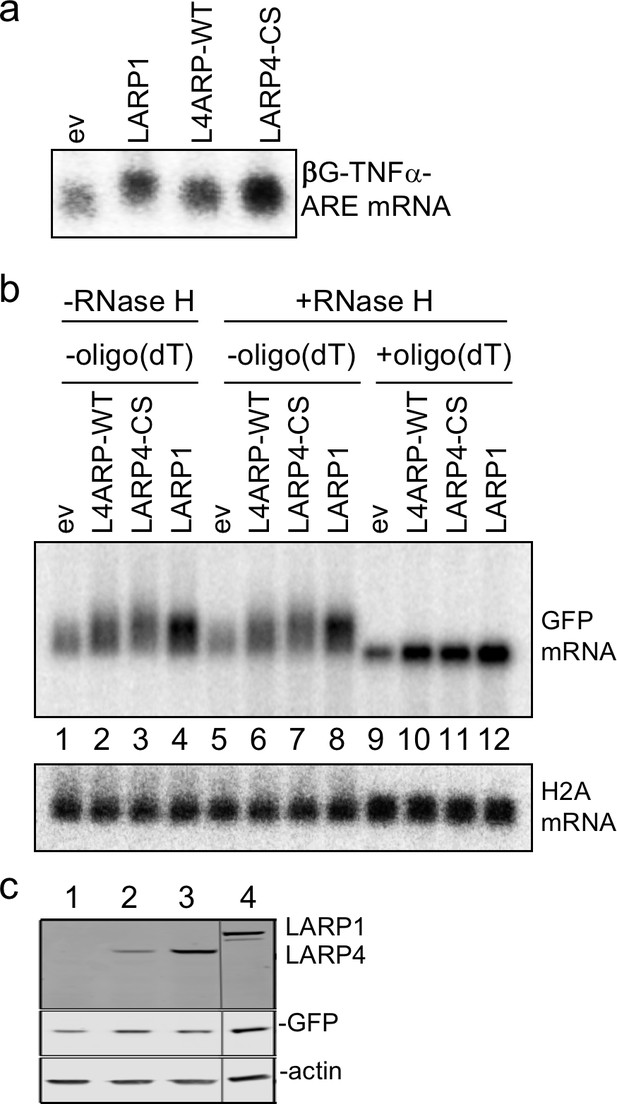
LARP1 promotes PAT 3' length stabilization of non-5'TOP mRNAs.
(a) Northern blot after transfection of HEK293 cells with expression plasmids indicated above the lanes, and the βG-TNFα-ARE reporter. (b) Same as in a but transfected with GFP followed by RNase H assay in presence or absence of oligo(dT) probed for GFP mRNA as indicated. The blot in b was probed for histone H2A. (c) Western blot showing LARP proteins expressed.
Tables
tRNA read counts.
https://doi.org/10.7554/eLife.28889.008| tRNA # | Codon(s) | Anticodon | AA | AA | Read count | Total bin | Fxn total | |
|---|---|---|---|---|---|---|---|---|
| 1 | ACA/G/C | TGT | T | Thr | 6484 | |||
| 2 | CCU/C | AGG | P | Pro | 6619 | |||
| 3 | ACG | CGT | T | Thr | 7664 | |||
| 4 | CCG | CGG | P | Pro | 9928 | |||
| 5 | AUA | TAT | I | Ile | 11902 | |||
| 6 | CAA | TTG | Q | Gln | 12533 | |||
| 7 | CCA/G/U | TGG | P | Pro | 12777 | |||
| 8 | AUU/C | AAT | I | Ile | 15657 | |||
| 9 | ACU/C | AGT | T | Thr | 16261 | |||
| 10 | AGC/U | GCT | S | Ser | 19618 | |||
| 11 | UUU/C | GAA | F | Phe | 20453 | total bin1 | 139896 | 0.05387 |
| 12 | UUA | TAA | L | Leu | 23330 | |||
| 13 | CUA | TAG | L | Leu | 23813 | |||
| 14 | AGG | CCT | R | Arg | 24943 | |||
| 15 | GGG | CCC | G | Gly | 25067 | |||
| 16 | GUA | TAC | V | Val | 25169 | |||
| 17 | UCG | CGA | S | Ser | 25390 | |||
| 18 | CAU/C | GTG | H | His | 25607 | |||
| 19 | GCG | CGC | A | Ala | 25778 | |||
| 20 | CUU/C | AAG | L | Leu | 26991 | |||
| 21 | UGG | CCA | W | Trp | 30456 | |||
| 22 | UUG | CAA | L | Leu | 31930 | total bin2 | 288474 | 0.11109 |
| 23 | CAG | CTG | Q | Gln | 33162 | |||
| 24 | UGU/C | GCA | C | Cys | 34487 | |||
| 25 | CGG | CCG | R | Arg | 35475 | |||
| 26 | GGA | TCC | G | Gly | 36754 | |||
| 27 | CGA | TCG | R | Arg | 40154 | |||
| 28 | UCA | TGA | S | Ser | 40530 | |||
| 29 | UCU/C | AGA | S | Ser | 41241 | |||
| 30 | AGA | TCT | R | Arg | 52949 | |||
| 31 | GAA | TTC | E | Glu | 55795 | |||
| 32 | GAU/C | GTC | D | Asp | 57771 | |||
| 33 | CUG | CAG | L | Leu | 61112 | |||
| 34 | CGU/C | ACG | R | Arg | 66635 | total bin3 | 556065 | 0.21414 |
| 35 | GCU/C | AGC | A | Ala | 74964 | |||
| 36 | GGU/C | GCC | G | Gly | 77978 | |||
| 37 | GUU/C | AAC | V | Val | 85733 | |||
| 38 | GCA | TGC | A | Ala | 92573 | |||
| 39 | AUG | CAT | M | Met | 102919 | |||
| 40 | GAG | CTC | E | Glu | 113500 | |||
| 41 | GUG | CAC | V | Val | 140741 | |||
| 42 | AAA | TTT | K | Lys | 161366 | |||
| 43 | UAU/C | GTA | Y | Tyr | 181682 | |||
| 44 | AAU/C | GTT | N | Asn | 239876 | |||
| 45 | AAG | CTT | K | Lys | 340378 | total bin4 | 1611710 | 0.62068 |
| Total | 2596700 |
ctAI scores for the CRD regions of LARP4 constructs (top) as well as the corresponding full length LARP4 constructs and some other reference proteins (bottom).
https://doi.org/10.7554/eLife.28889.009| CRD: | ctAI | Comments |
|---|---|---|
| WT | 0.3099 | |
| CSc | 0.3444 | Differs from WT by 13 synonymous substitutions that require decoding by different tRNA anticodons. |
| CS-I | 0.3802 | |
| CS-R | 0.4157 | |
| CS-B | 0.4638 | |
| CS-Tyr | 0.5082 | Differs from CS-B by U-to-C wobble substitutions at all 7 Tyr codons. |
| CSb | 0.3722 | Differs from WT by 14 U-to-C wobble substitutions (see text) but not Asn codons |
| CSa | 0.3973 | Differs from CSb only by U-to-C wobble substitutions at 5 Asn codons. |
| CS-W | 0.3028 | |
| Full length: | ctAI | |
| LARP4-WT | 0.4200 | |
| LARP4-CS-B | 0.4370 | |
| LARP4-CS-Tyr | 0.4409 | |
| LARP4-CS-W | 0.4190 | |
| LARP4B | 0.4286 | |
| LARP4B with LARP4 CS-B CRD | 0.4392 | |
| LARP4B with LARP4 CS-Tyr CRD | 0.4431 | |
| LARP4B with L4 WT CRD | 0.4227 | |
| hRpl35 | 0.6020 | |
| hActin (ACTG1) | 0.4883 | |
| hRps28 | 0.5190 | |
| eGFP | 0.5368 | |
| GAPDH | 0.4531 | |
| hRpl32 | 0.4866 | |
| H2A | 0.5347 |
Additional files
-
Supplementary file 1
Table list of s oligonucleotide probe sequences and their hybridization incubation temperatures (Ti).
- https://doi.org/10.7554/eLife.28889.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28889.018





