Heg1 and Ccm1/2 proteins control endocardial mechanosensitivity during zebrafish valvulogenesis
Figures
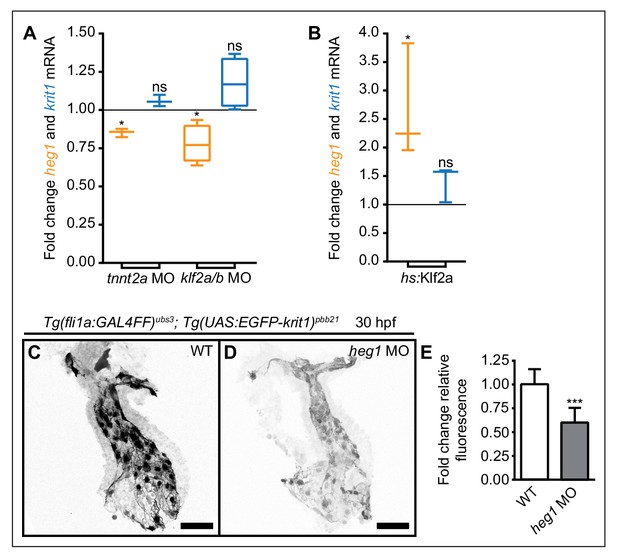
Mechanosensitive expression of heg1 mRNA and stabilization of Krit1 protein levels by heg1.
(A) RT-qPCR quantifications of heg1 and krit1 mRNA levels at 54 hpf in morphant embryos lacking blood-flow [troponin T type 2a (tnnt2a)], or Krüppel like factors 2a and 2b (klf2a/b) compared with wild-type expression levels. (B) RT-qPCR quantification showing that heat-shock-induced overexpression of klf2a causes significant upregulation of heg1 but not of krit1 expression at 54 hpf. (C–E) Morpholino-mediated knockdown of Heg1 significantly reduces endocardial EGFP-Krit1 protein levels of 30 hpf Tg(fli:GAL4FF)ubs3; Tg(UAS:EGFP-Krit1)pbb21 embryos. Scale bars are 50 µm. Mean values ± SEM of three (tnnt2a MO, hs:klf2a, heg1 MO) or four (klf2a/b MO) individual experiments are shown. Ratio paired (A, B) or unpaired (E) t-test was used to compare each condition with controls (ns: not significant, *p<0.05, ***p<0.001).
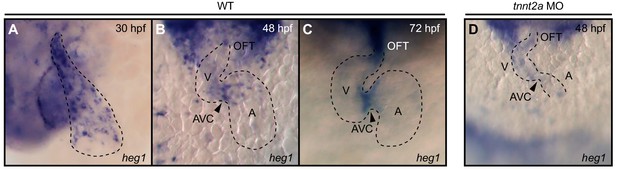
Whole-mount in situ hybridization of heg1 cardiac expression.
(A) At 30 hpf, heg1 is expressed throughout the entire endocardium. (B) Later, at 48 hpf, heg1 expression becomes restricted to high fluid shear stress regions of the endocardium including the atrioventricular canal (AVC). (C) The strong expression of heg1 at the AVC persists at 72 hpf. (D) Expression of heg1 was reduced in morphant embryos lacking blood-flow [troponin T type 2a (tnnt2a)] at 48 hpf. A: atrium, V: ventricle, OFT: outflow tract, arrowheads in B, C, and D point at the AVC.
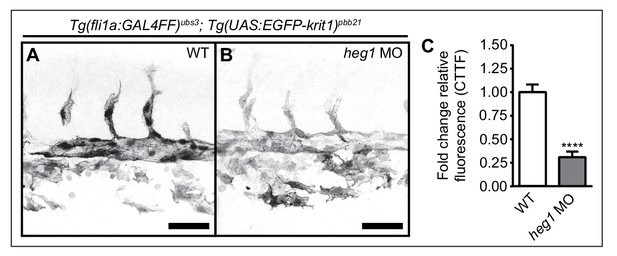
Stabilization of Krit1 protein levels by heg1 in the trunk vasculature.
(A–C) Morpholino-mediated knockdown of Heg1 significantly reduces EGFP-Krit1 protein levels in the trunk vasculature of 30 hpf Tg(fli:GAL4FF)ubs3; Tg(UAS:EGFP-Krit1)pbb21 embryos. Scale bars are 50 µm. Mean values ± SEM of three individual experiments are shown. Unpaired t-test was used to compare conditions with control (****p<0.0001).
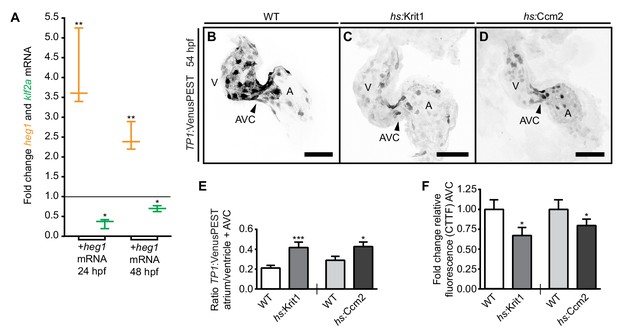
Desensitization of endocardial cells to blood-flow by heg1, krit1, or ccm2 overexpression.
(A) Correlation of high heg1 and low klf2a mRNA expression levels at 24 and 48 hpf as determined by RT-qPCR. (B–D) Projections of confocal z-stack images of 54 hpf hearts expressing the flow-responsive marker Tg(TP1:VenusPEST)s940 in heat-shocked wild-type (WT) (B), heat-shock- (hs) induced krit1 overexpression (C), and hs-induced ccm2 overexpression (D). (E) Different ratios of corrected total tissue fluorescence (CTTF) of atrial versus ventricular (+atrioventricular canal region (AVC), arrowhead) expression of the flow-responsive marker Tg(TP1:VenusPEST)s940 between 54 hpf WT and krit1-overexpressing hearts (n = 37) and WT and ccm2-overexpressing hearts (n = 39). krit1- or ccm2-overexpression results in more equal chamber expression of flow-responsive marker Tg(TP1:VenusPEST)s940 expression (depicted is a WT heart corresponding to the krit1-overexpression experiment). (F) Fold change of relative fluorescence (CTTF) in the AVC of Tg(TP1:VenusPEST)s940 in krit1- and ccm2- overexpressing hearts versus the corresponding controls. Scale bars are 50 µm. Mean values ± SEM are shown. Unpaired t-test was used to compare each condition with the WT (*p<0.05, **p<0.01, ***p<0.001).
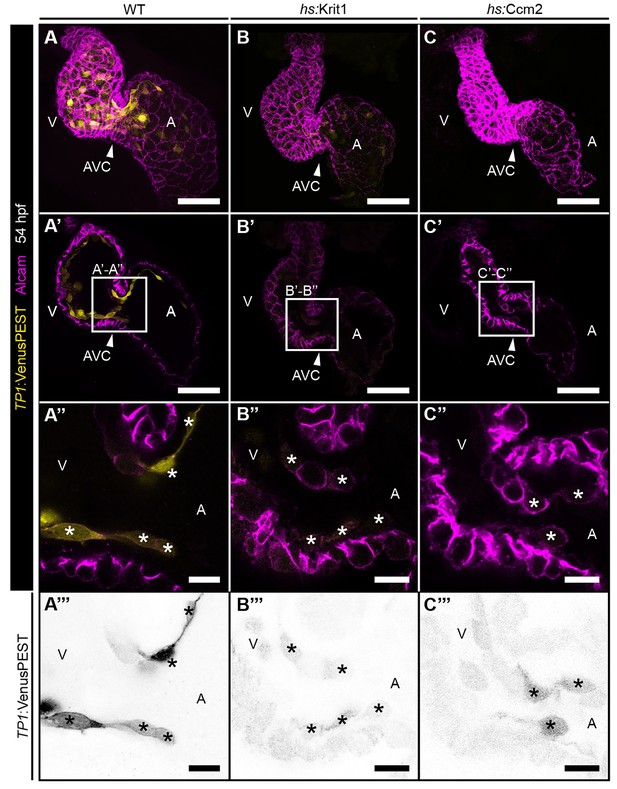
Exclusive endocardial expression of the Notch reporter Tg(TP1:VenusPEST)s940 and cardiac cushions forming normally on overexpression of krit1 and ccm2.
(A–C) (A) Maximum projections of confocal z-stacks of 48 hpf heat-shocked wild-type (WT) embryo, (B) heat-shock-induced (hs) krit1 overexpressing embryo, and (C) hs-induced ccm2 overexpressing embryo. Myocardial cells and endocardial cushions are stained for Alcam (Magenta). Myocardial cells do not express the destabilized Notch reporter Tg(TP1:VenusPEST)s940 (yellow), which is expressed within endocardium and endocardial cushions. (A’–C’) Shown are projections of 3 confocal z-stack planes (1 μm each) of the same hearts as shown in (A–C). (A’’–C”’) Single confocal z-section images of the atrioventricular canal (AVC) region highlighted with a white square in the panels D-F. (A’’’–C’’’) Inverted images of Notch reporter shown in A’’-C’’ (asterisks mark endocardial cushion cells). Note dowregulation of Notch activity on Krit1 and Ccm2 overexpression (B, C, B’–C’’’). The morphology of endocardial cushions remains unaltered on krit1 and ccm2 overexpression at this stage (B’’–C’’). A: atrium, V: ventricle, arrowheads point at the AVC. Scale bars are 50 µm in A-F and 10 µm in D’-F’’.
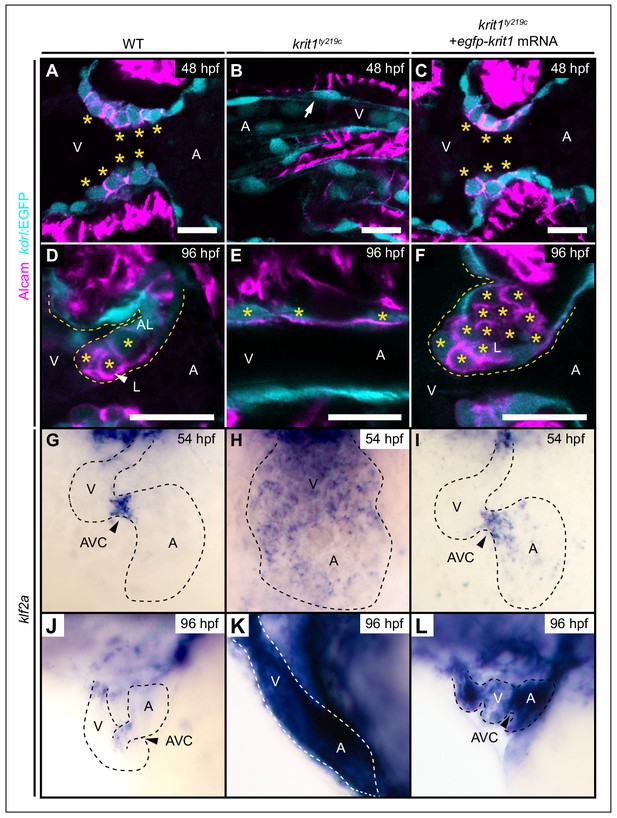
Krit1 is required for abluminal cell fates during valvulogenesis.
(A–F) Single confocal z-section images of endocardial cells within the atrioventricular canal (AVC) region marked by Tg(kdrl:EGFP)s843 expression (cyan) and Alcam staining (magenta). (A) At 48 hpf, wild-type (WT) endocardial cushion cells express Alcam (asterisks). (B) In krit1ty219c mutants, endocardial cells of the AVC region do not express Alcam and cushions do not form (arrow). (C) Injection of egfp-krit1 mRNA rescues cardiac cushion formation in krit1ty219c mutants (n = 15/15). krit1ty219c mutant hearts form cardiac cushions and endocardial cushion cells express Alcam. (D) By 96 hpf, Alcam expression is mainly restricted to luminal endocardial cells of the developing WT cardiac valve leaflet (asterisks). (E) krit1ty219c mutants do not form valve leaflets but AVC endocardial cells express Alcam (asterisks). (F) krit1ty219c mutant that was initially rescued by egfp-krit1 mRNA injection has a dysmorphic cardiac valve leaflet at 96 hpf with an agglomeration of luminal Alcam-positive cells (asterisks). (G–L) Whole-mount in situ hybridization of klf2a cardiac expression. (G) At 54 hpf, klf2a expression is restricted to endocardial cells of the AVC in WT while (H) klf2a is strongly expressed throughout the entire endocardium in krit1ty219c mutant. (I) krit1ty219c mutant rescued by injection of egfp-krit1 mRNA has a restricted and strong klf2a expression at the AVC (n = 6/6). (J) By 96 hpf, klf2a expression is restricted to the AVC in WT while (K) klf2a is strongly expressed throughout the entire endocardium in krit1ty219c mutants. (L) krit1ty219c mutant embryo injected with egfp-krit1 mRNA has high klf2a levels throughout the entire endocardium (n = 8/8). A: atrium, V: ventricle, L: luminal, AL: abluminal. Scale bars are 10 μm.
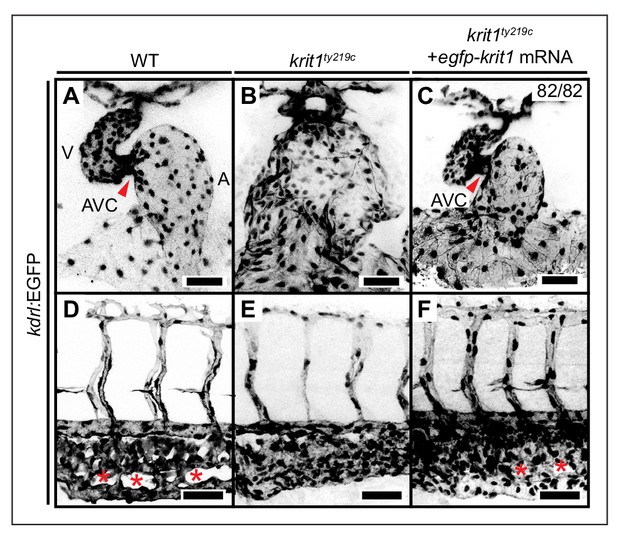
Rescue of krit1ty219c mutant cardiovascular phenotypes by injection of mRNA encoding EGPF-Krit1.
(A–C) Projections of confocal z-stack images of (A) 48 hpf wild-type (WT), (B) krit1ty219c mutant, and (C) rescued krit1ty219c mutant hearts marked by Tg(kdrl:EGFP)s843 expression. In comparison to WT, krit1ty219c mutant hearts are dilated and lack the atrioventricular canal (AVC) (red arrowhead). (C) Injection of egfp-krit1 mRNA into krit1ty219c mutants rescues the krit1ty219c mutant cardiac phenotype (n = 82/82 krit1ty219c mutant embryos rescued). (D–F) Projections of confocal z-stack images of 48 hpf caudal plexus regions of the vasculature marked by Tg(kdrl:EGFP)s843 expression. (D) In comparison to the highly branched WT morphology of the caudal vein plexus (red asterisks), (E) the krit1ty219c mutant vasculature has a fused morphology. (F) Injection of egfp-krit1 mRNA into krit1ty219c mutants rescues the krit1ty219c mutant vascular phenotype including a highly branched WT-like caudal vein plexus (red asterisks). A: atrium, V: ventricle. Scale bars are 50 µm.
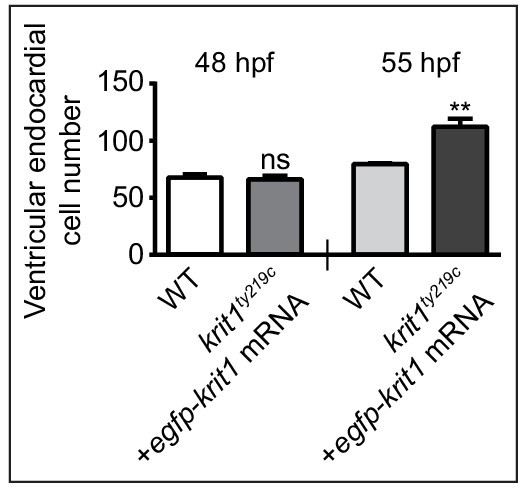
Injection of mRNA encoding EGFP-Krit1 into krit1ty219c mutants prevents overgrowth of ventricular endocardial cells.
Quantification of ventricular endocardial cell numbers reveals normal numbers in rescued krit1ty219c mutants by 48 hpf. At 55 hpf, ventricular endocardial cell numbers in krit1ty219c mutants injected with mRNA encoding EGFP-Krit1 increase considerably compared to wild-type (WT). Mean values ± SEM are shown. ns: not significant, *p<0.05, **p<0.01.
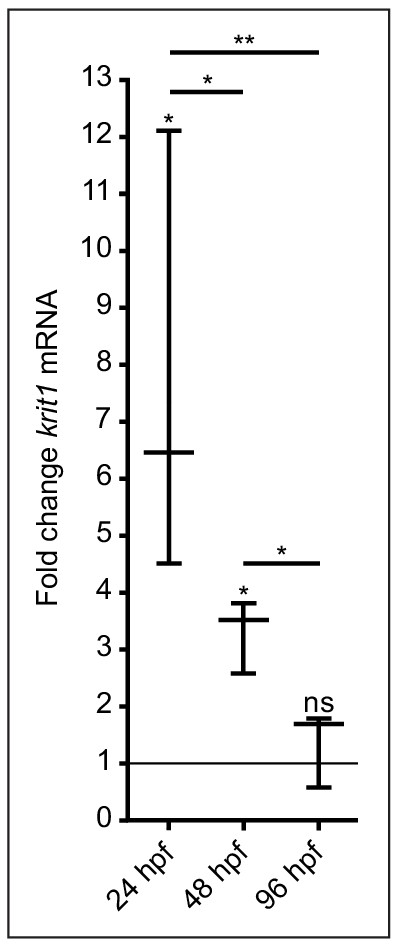
krit1 expression decreases to a basal level by 96 hpf in rescued embryos.
Significant decrease of krit1 levels from 24 to 48 hpf and from 48 to 96 hpf as determined by RT-qPCR. Mean values ± SEM are shown. ns: not significant, *p<0.05, **p<0.01.
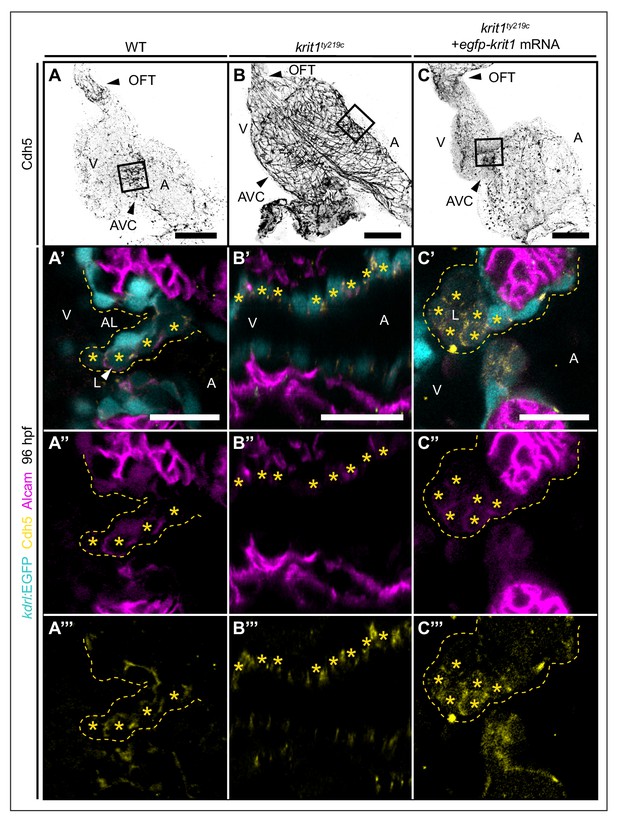
Cadherin-5 is misexpressed in krit1ty219c mutant embryonic hearts.
(A–C) Projections of confocal z-stack images of 96 hpf embryonic hearts marked by Cadherin 5 (Cdh5) staining. (A’–C’’’) Details are shown in single confocal plane sections of the atrioventricular canal (AVC) region (boxes) of superior cardiac valve leaflets of 96 hpf hearts marked by Tg(kdrl:EGFP)s843 expression (cyan), Cdh5 staining (yellow), and Alcam staining (magenta). (A’–A’’’) At 96 hpf, Alcam and Cdh5 expression is mainly restricted to luminal endocardial cells of the developing wild-type (WT) cardiac valve leaflet (asterisks). (B’–B’’’) krit1ty219c mutants do not form valve leaflets and endocardial cells express Alcam and Cdh5 (asterisks). (C’–C’’’) Injection of egfp-krit1 mRNA into krit1ty219c mutants results in a dysmorphic cardiac valve leaflet at 96 hpf with an agglomeration of luminal leaflet cells marked by Alcam and Cdh5 expression (asterisks) (n = 17/23 krit1ty219c mutants). A: atrium, V: ventricle, L: luminal, AL: abluminal. Scale bars are 50 μm (A–C) and 20 µm (A’–C’’’).
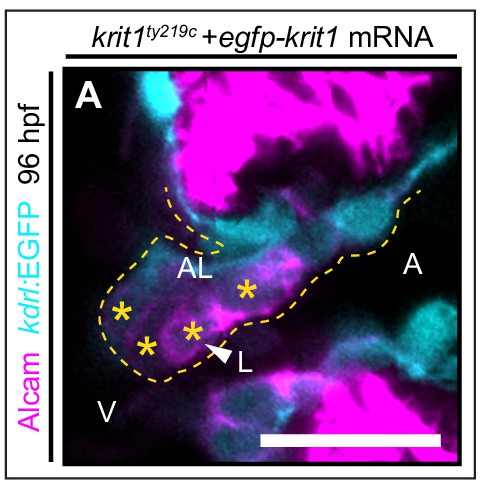
Krit1 is required for abluminal cell fates during cardiac valve leafet formation.
(A) Details of a single confocal plane section of the atrioventricular canal (AVC) region of superior cardiac valve leaflet. A group of krit1ty219c mutants injected with egfp-krit1 mRNA showing a normal distribution of Alcam (magenta) positive luminal endocardial cells by 96 hpf (n = 6/23 krit1ty219c mutants). A: atrium, V: ventricle, L: luminal, AL: abluminal. Scale bars are 10 µm.
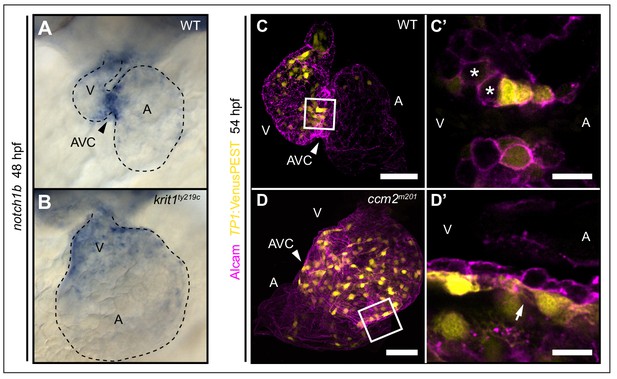
Notch activity within endocardial cushion cells is affected in ccm mutants.
(A, B) Whole-mount in situ hybridization of notch1b cardiac expression at 48 hpf. (A) notch1b expression is restricted to endocardial cells of the atrioventricular canal (AVC) in wild-type (WT), while notch1b is strongly expressed throughout all ventricular cells in krit1ty219c mutants (B). (C, D) Projections of single confocal z-section images of endocardial cells marked by Tg(TP1:VenusPEST)s940 expression (yellow) and Alcam staining (magenta) at 54 hpf. (C) In WT, Notch activity is highest in endocardial cells of the AVC and outflow regions. (C’) Single confocal plane section of the AVC (white box in C) reveals that some endocardial cells close to the ventricle lack Notch activity (asterisks). (D) In ccm2m201 mutants, the domain of high Notch activity is expanded to most ventricular endocardial cells. (D’) Single confocal plane section of the AVC region (box in D, arrow) shows high Notch expression in all endocardial cells of the AVC region. A: atrium, V: ventricle, Scale bars are 50 μm (C, D), and 10 µm (C’, D’).
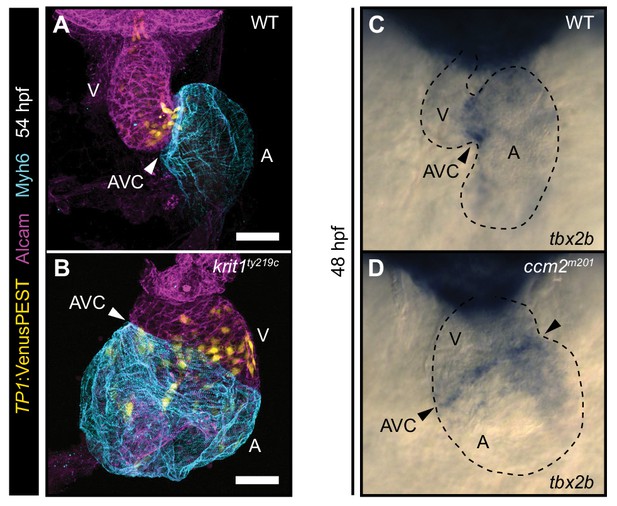
The region of Notch activity within the endocardium is expanded in krit1ty219c mutants.
(A, B) Projections of confocal z-stack images of embryonic hearts marked by Tg(TP1:VenusPEST)s940 expression (yellow), Myh6 staining (cyan), and Actin staining (magenta) at 54 hpf. (A) In wild-type (WT), Notch activity is highest in endocardial cells of the atrioventricular canal (AVC) and outflow regions. (B) In krit1ty219c mutants, the domain of high Notch activity is expanded to ventricular and atrial endocardial cells. (C, D) Whole-mount in situ hybridization of tbx2 cardiac expression at 48 hpf. (A) Tbx2 expression is restricted to the atrioventricular canal (AVC) both in wild-type (WT) and in ccm2m201 mutants (B). Arrowheads point at the AVC. A: atrium, V: ventricle, Scale bars are 50 μm (A, B).
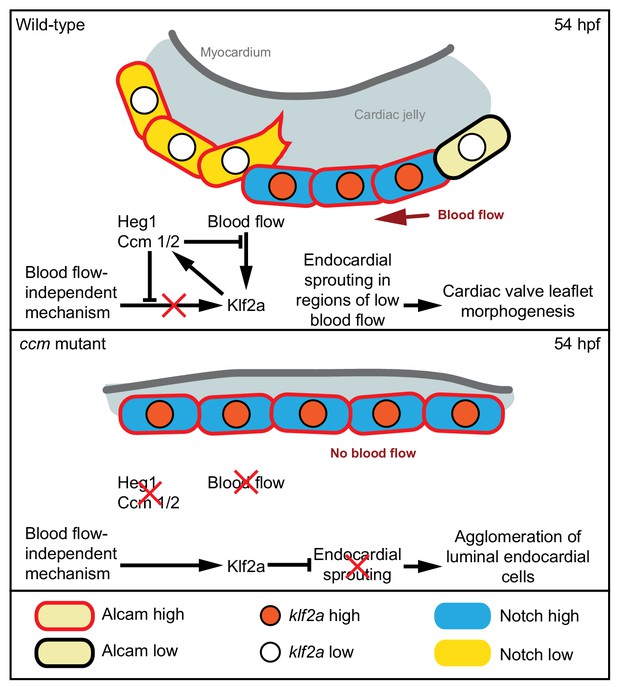
Model of the molecular pathways involved in endocardial cushion cell sprouting at 54 hpf.
In wild-type, endocardial cells exposed to blood-flow have high levels of Klf2a, Notch, Alcam, and maintain cell adhesion. Ccm proteins provide a bias in Klf2a and Notch expression levels. Some cells that express lower levels of klf2a and have lower Notch activity establish protrusions and migrate into the cardiac jelly (endocardial sprouting). In ccm mutants, endocardial cells overexpress klf2a and have higher Notch activity because of a blood-flow-independent mechanism. As a consequence, high levels of Notch activity throughout the entire endocardium result in failure of endocardial sprouting.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene (heg1) | heg1 | ZFIN | ZFIN ID: ZDB-GENE-040714–1 | |
| gene (krit1) | krit1 | ZFIN | ZFIN ID: ZDB-GENE-030131–555 | |
| gene (klf2a) | klf2a | ZFIN | ZFIN ID: ZDB-GENE-011109–1 | |
| gene (klf2b) | klf2b | ZFIN | ZFIN ID: ZDB-GENE-011109–2 | |
| gene (tnnt2a) | tnnt2a | ZFIN | ZFIN ID: ZDB-GENE-000626–1 | |
| gene (ccm2) | ccm2 | ZFIN | ZFIN ID: ZDB-GENE-040712–6 | |
| gene (notch1b) | notch1b | ZFIN | ZFIN ID: ZDB-GENE-990415–183 | |
| strain, strain background (krit1ty219c) | krit1ty219c | DOI:10.1242/dev.02469 | ZFIN ID: ZDB-ALT-980203–1289 | |
| strain, strain background (ccm2m201) | ccm2m201 | PMID:9007227 | ZFIN ID: ZDB-ALT-980203–523 | |
| strain, strain background (Tg(EPV.Tp1-Mmu.Hbb: Venus-Mmu.Odc1)s940) | Tg(EPV.Tp1-Mmu.Hbb: Venus-Mmu.Odc1)s940 | DOI:10.1242/dev.076000 | ZFIN ID: ZDB-ALT-120419–6 | |
| strain, strain background (Tg(kdrl:EGFP)s843) | Tg(kdrl:EGFP)s843 | DOI:10.1242/dev.02087 | ZFIN ID: ZDB-ALT-050916–14 | |
| strain, strain background (Tg(fli1a:GAL4FF)ubs3) | Tg(fli1a:GAL4FF)ubs3 | DOI:10.1016/j.cub. 2011.10.016 | ZFIN ID: ZDB-ALT-120113–6 | |
| strain, strain background (Tg(hsp70l:Krit1_IRES_EGFP)md6) | Tg(hsp70l: Krit1_IRES_EGFP)md6 | this paper | ||
| strain, strain background (Tg(hsp70l:Ccm2_IRES_EGFP)md12) | Tg(hsp70l: Ccm2_IRES_EGFP)md12 | this paper | ||
| strain, strain background (Tg(hsp70l:Klf2a_IRES_EGFP)pbb22) | Tg(hsp70l: Klf2a_IRES_EGFP)pbb22 | this paper | ||
| strain, strain background (Tg(UAS:EGFP-Krit1, cryaa:EGFP)pbb21) | Tg(UAS:EGFP-Krit1, cryaa:EGFP)pbb21 | this paper | ||
| genetic reagent (tnnt2a morpholino) | tnnt2a MO | DOI:10.1038/ng875 | ZFIN ID: ZDB-MRPHLNO-060317–4 | |
| genetic reagent (klf2a morpholino) | klf2a MO | DOI:10.1038/nature08889 | ZFIN ID: ZDB-MRPHLNO-100610–9 | 2 ng/embryo |
| genetic reagent (klf2b morpholino) | klf2b MO | DOI:10.1016/j.devcel. 2014.12.016 | ZFIN ID: ZDB-MRPHLNO-150427–1 | 1 ng/embryo |
| genetic reagent (heg1 morpholino) | heg1 MO | PMID:14680629 | ZFIN ID: ZDB-MRPHLNO-080714–5 | 5 ng/embryo |
| genetic reagent (klf2a probe) | klf2a | DOI:10.1016/j.devcel. 2014.12.016 | ZFIN ID: ZDB-FIG-150407–1 | 5 ng/embryo |
| genetic reagent (notch1b probe) | notch1b | DOI:10.1126/science. 293.5535.1670 | ZFIN ID: ZDB-FIG-151113–25 | |
| genetic reagent (heg1 probe) | heg1 | DOI: 10.1242/dev.143362 | ZFIN ID: ZDB-PUB-031217–1 | |
| genetic reagent (tbx2 probe) | tbx2 | DOI: 10.1002/dvdy.22622 | ZFIN ID: ZDB-PUB-110502–3 | |
| Antibody (rabbit anti-VE- Cadherin (Cdh5)) | Cdh5 | DOI:10.1016/j.ydbio. 2008.01.038 | ZFIN ID: ZDB-PUB-080326–18 | 1:200 |
| Antibody (mouse anti-Zn-8/Alcam) | Alcam | Developmental Studies Hybridoma Bank | ZFIN ID: ZDB-ATB-081002–22 | 1:25 |
| Antibody (mouse anti-Myh6) | Myh6 | Developmental Studies Hybridoma Bank | ZFIN ID: ZDB-ATB-081002–54 | 1:10 |
| Antibody (Alexa Fluor 633-conjugated goat anti-rabbit) | Alexa Fluor 633- conjugated goat anti-rabbit | Invitrogen A21070 | 1:200 | |
| Antibody (Rhodamine Red- X-conjugated goat anti-mouse) | Rhodamine Red- X- conjugated goat anti-mouse | Jackson ImmunoResearch Laboratories 115-295-003 | 1:200 | |
| Antibody (Dylight 649- conjugated goat anti-mouse) | Dylight 649- conjugated goat anti-mouse | Jackson ImmunoResearch Laboratories 115-495-003 | 1:200 | |
| recombinant DNA reagent (pDestTol2 (#426)) | pDestTol2 | N. Lawson | Lawson Lab: #426 | |
| recombinant DNA reagent (p5E-hsp70l (#222)) | p5E-hsp70l | N. Lawson | Lawson Lab: #222 | |
| recombinant DNA reagent (p5E-CMV/SP6 (#382)) | p5E-CMV/SP6 | Chien, Univ. Utah | Tol2kit: #382 | |
| recombinant DNA reagent (p5E-UAS (#327)) | p5E-UAS | Chien, Univ. Utah | Tol2kit: #327 | |
| recombinant DNA reagent (p3E-IRES_EGFPpA (#389)) | p3E-IRES_EGFPpA | N. Lawson | Lawson Lab: #389 | |
| recombinant DNA reagent (p3E-cryaa:EGFPpA) | p3E-cryaa:EGFPpA | other | plasmid was generated in our lab | |
| recombinant DNA reagent (p3E-EGFPpA (#366)) | p3E-EGFPpA | Chien, Univ. Utah | Tol2kit: #366 | |
| recombinant DNA reagent (p3E-pA (#383)) | p3E-pA | Chien, Univ. Utah | Tol2kit: #383 | |
| recombinant DNA reagent (pDest_Tol2pA_Hsp70l: Krit1_IRES_EGFPpA) | pDest_Tol2pA_Hsp70l: Krit1_IRES_EGFPpA | this paper | ||
| recombinant DNA reagent (pDest_Tol2pA_Hsp70l: Ccm2_IRES_EGFPpA) | pDest_Tol2pA_Hsp70l: Ccm2_IRES_EGFPpA | this paper | ||
| recombinant DNA reagent (p3E-pA (pDest_Tol2pA_ Hsp70l:Klf2a_IRES_EGFPpA) | pDest_Tol2pA_Hsp70l: Klf2a_IRES_EGFPpA | this paper | ||
| recombinant DNA reagent (pDest_Tol2pA_UAS: EGFP-Krit1pA,cryaa:EGFPpA) | pDest_Tol2pA_UAS: EGFP-Krit1pA,cryaa:EGFPpA | this paper | ||
| recombinant DNA reagent (pDest_Tol2pA_CMV/SP6: EGFP-Krit1pA) | pDest_Tol2pA_CMV/SP6: EGFP-Krit1pA | this paper | ||
| sequence-based reagent (qPCR-primer heg1 FW) | heg1 FW | this paper | ||
| sequence-based reagent (qPCR-primer heg1 RW) | heg1 RW | this paper | ||
| sequence-based reagent (qPCR-primer krit1 FW) | krit1 FW | this paper | ||
| sequence-based reagent (qPCR-primer krit1 RW) | krit1 RW | this paper | ||
| sequence-based reagent (qPCR-primer klf2a FW) | klf2a FW | this paper | ||
| sequence-based reagent (qPCR-primer klf2a RW) | klf2a RW | this paper | ||
| sequence-based reagent (qPCR-primer eif1b FW) | eif1b FW | this paper | ||
| sequence-based reagent (qPCR-primer eif1b RW) | eif1b RW | this paper | ||
| commercial assay or kit (SP6 polymerase (mMessage Machine kit, Ambion)) | SP6 polymerase | Ambion | Ambion:AM1340 | |
| commercial assay or kit (RevertAid H Minus First Strand cDNA Synthesis kit (ThermoFisher Scientific)) | RevertAid H Minus First Strand cDNA Synthesis kit | ThermoFisher Scientific | ThermoFisher Scientific:K1631 | |
| commercial assay or kit (KAPA Sybr Fast qPCR kit (Peqlab)) | KAPA Sybr Fast qPCR kit | Peglab | Peglab:4385612 | |
| chemical compound, drug (1- phenyl-2-thiourea (PTU)) | PTU | Sigma Aldrich | Sigma Aldrich:P7629 | 0.003% |
| chemical compound, drug (Rhodamine-Phalloidin) | Rhodamine-Phalloidin | Invitrogen | Invitrogen:R415 | 1:250 |
| chemical compound, drug (Tricaine (3-amino benzoic acidethylester)) | Tricaine | Sigma-Aldrich | Sigma Aldrich:A-5040 | 0.16 mg/ml |
| software, algorithm (GraphPad Prism6) | GraphPad Prism6 | GraphPad | ||
| software, algorithm (Imaris (Bitplane, Version 8.1)) | Imaris | Bitplane | ||
| software, algorithm (Fiji software) | Fiji | DOI:10.1038/nmeth.2019 | ||
| software, algorithm (Adobe Bridge and Photoshop (Adobe Systems)) | Adobe Bridge and Photoshop | Adobe | ||
| software, algorithm (Zen 8.1 Software (Zeiss)) | Zen | Zeiss | ||
| software, algorithm (Excel 2010 (Microsoft Office)) | Excel | Microsoft | ||
| software, algorithm (PikoReal software 2.2 (ThermoFisher Scientific)) | PikoReal software | ThermoFisher Scientific |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28939.016





