Ubiquitin turnover and endocytic trafficking in yeast are regulated by Ser57 phosphorylation of ubiquitin
Figures
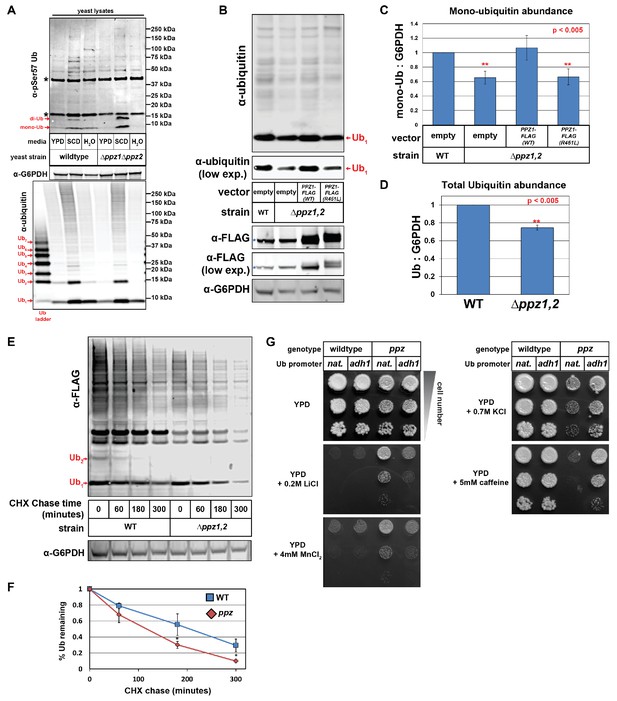
Ppz phosphatase activity is required for proper management of cellular ubiquitin.
(A) Immunoblot analysis using an α-phospho-Ser57 specific antibody to detect Ser57 phosphorylation of ubiquitin in yeast lysates. Yeast lysates from a SUB280 background, comparing wild-type cells (left three lanes) and ppz mutant cells (right three lanes) grown to mid log phase in YPD (‘YPD’), or shifted from mid log growth in YPD to minimal complete media for 6 hr (‘SCD’) or shifted to growth in water overnight (‘H2O’). Samples were resolved by SDS-PAGE and immunoblotted using antibodies that recognize total ubiquitin (bottom panel), G6PDH (middle panel), or phospho-Ser57 ubiquitin (top panel). For the total ubiquitin blot (bottom panel), a ubiquitin ladder was included as a standard for different unconjugated poly-ubiquitin species (red arrows). For the α–phospho-Ser57 ubiquitin blot (top panel), red arrows indicate species that are specific to Ser57 on ubiquitin and asterisks (*) indicate non-specific bands detected by the antibody, as illustrated in Figure 1—figure supplement 2. (B) The indicated yeast cells (SEY6210 background) containing either empty vector or PPZ1-FLAG vectors were analyzed for total cellular ubiquitin levels by immunoblot analysis. The blue asterisk indicates a background band detectable by FLAG antibody that is a MW similar to Ppz1-FLAG. (C) Quantification of mono-ubiquitin abundance in yeast cell lysates (SEY6210 background) from multiple biological replicates of the immunoblot shown in (B) (n = 5). (D) Total ubiquitin abundance was quantified in wild-type and ppz mutant cells expressing endogenously FLAG-tagged ubiquitin. Total cell lysates were analyzed by quantitative analysis of slot blots shown in Figure 1—figure supplement 3. (E) The indicated yeast cells were grown to mid-log phase and cell lysates were analyzed for total ubiquitin levels at the indicated time points following a cycloheximide (CHX) chase. (F) The results for (F) were quantified over multiple experiments (n = 3). (G) Analysis of yeast growth in the indicated conditions. In this experiment, the yeast ubiquitin gene was expressed from either native (pRPS31) or overexpression (pADH1) promoter was shuffled into the SUB280 strain background. Indicated yeast were plated in 10-fold serial dilutions on indicated plates. In all panels, double asterisk (**) indicates p<0.005 and single asterisk (*) indicates p<0.05.
-
Figure 1—source data 1
Results from SILAC-based quantitative comparison of the yeast phosphoproteome from wild-type (heavy) and Δppz1Δppz2 (light) cells.
Phosphorylation events that are elevated threefold or greater in Δppz1Δppz2 cells are shown. Also shown are phosphopeptides detected from Ppz1, which are present in wild-type cells and missing from Δppz1Δppz2 cells.
- https://doi.org/10.7554/eLife.29176.008
-
Figure 1—source data 2
This spreadsheet contains the quantification and statistical analysis for mono-ubiquitin levels (Figure 1C), total ubiquitin levels (Figure 1D), and for ubiquitin degradation in a cycloheximide chase experiment (Figure 1F).
- https://doi.org/10.7554/eLife.29176.009

A SILAC-based quantitative comparison of the phosphoproteome in wildtype (heavy) and Δppz1Δppz2 (light) cells.
The schematic on the left shows the experimental approach for quantitative analysis of the phosphoproteome in ppz mutant cells. The scatter plot on the right summarizes SILAC-based quantitative analysis of yeast phosphoproteomes comparing wildtype (heavy) and ppz mutant (light) cells. All phosphorylated peptides with a fold difference greater than three are colored red. Results of this analysis are tabulated in Figure 1—source data 1.

Analysis of antibodies recognizing pSer57 ubiquitin.
The left panel shows immunoblot analysis of synthetic ubiquitin, phospho-Ser57 ubiquitin, and phospho-Ser65 ubiquitin using an α-phospho-Ser57-specific antibody. The right panel shows immunblot analysis using the same antibody to probe lysates from yeast expressing only wild-type ubiquitin or Ser57Asp ubiquitin. Many background bands are apparent but two low MW bands corresponding to mono- and di-ubiquitin are recognized by the antibody in a Ser57-specific manner.
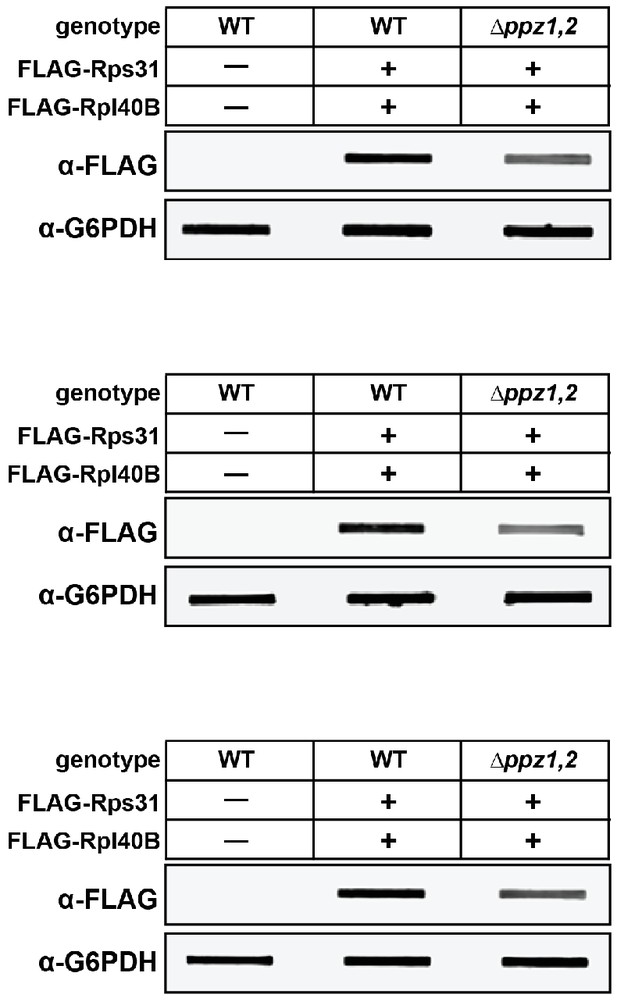
Quantification of cellular ubiquitin levels by analysis of slot blots.
Yeast strains (SEY6210 background) with endogenously FLAG-tagged ubiquitin (at the RPL40B and RPS31 loci with native promoter and terminator containing an N-terminal 3xFLAG tag) were grown to mid-log phase and precipitated in 10% cold TCA. Total cell lysates from solubilized pellets were bound to PVDF membranes using a vacuum manifold and immunoblotting analysis was performed as indicated. Three biological replicate experiments are shown.
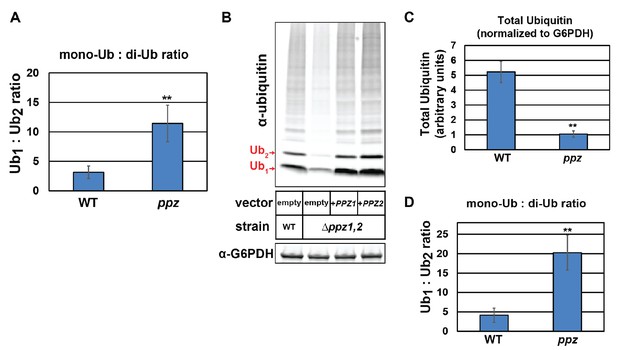
Characterization of ubiquitin levels and distribution in wild-type and Δppz1Δppz2 cells.
(A) Using quantitative immunoblots mono-ubiquitin: di-ubiquitin ratios were measured in yeast lysates (SEY6210 background) and averaged over multiple experiments (n = 5). (B) The indicated yeast cells (SUB280 background) containing either empty vector or native PPZ1 or PPZ2 complementation vectors were analyzed for total cellular ubiquitin levels by immunoblot analysis. (C) Analysis of total ubiquitin in SUB280 yeast lysates from cells grown to mid-log phase in YPD. (D) Analysis of mono-ubiquitin: di-ubiquitin ratios were measured in yeast lysates (SUB280 background) and averaged over multiple experiments (n = 5).

Catalytic activity of the Ppz1 phosphatase is required for phenotype complementation.
In this experiment, yeast cells (SEY6210 background) containing either empty vector of the indicated complementation vector were plated in 10-fold serial dilutions on indicated plates.
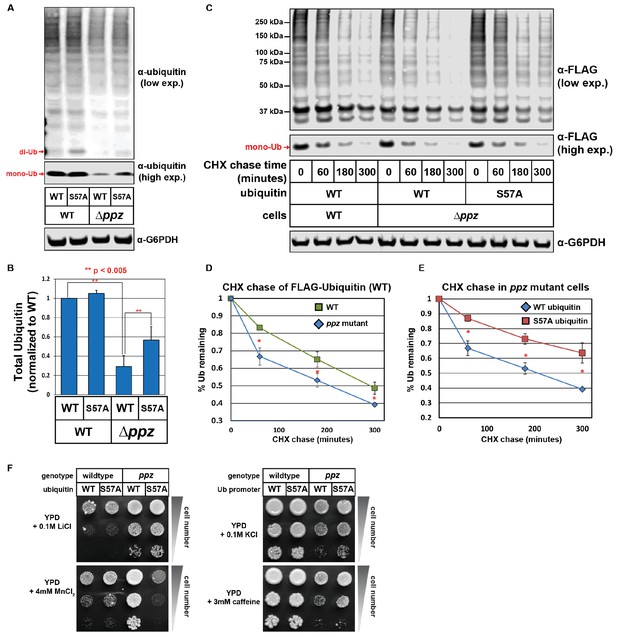
Ser57 phosphorylation of ubiquitin contributes to ppz mutant phenotypes.
(A) Quantitative immunoblot analysis was performed on lysates from wild-type or ppz mutant yeast cells (SUB280 background) expressing either wild-type ubiquitin or Ser57Ala ubiquitin. (B) The results for (A) were quantified over multiple experiments (n = 3). (C) Quantitative immunoblot analysis was performed on lysates from wild-type or ppz mutant yeast cells (SUB280 background) expressing either wild-type ubiquitin or Ser57Ala FLAG-ubiquitin (driven by the TEF1 promoter) following addition of cycloheximide (CHX). (D and E) The results for (C) were quantified over multiple experiments (n = 3). (F) Wild-type and ppz mutant yeast strains (SUB280 background) expressing only wild-type or Ser57Ala ubiquitin were plated in 10-fold serial dilution on the indicated media. Plates were imaged after three days of growth at 26°C. In all panels, double asterisk (**) indicates p<0.005 and single asterisk (*) indicates p<0.05.
-
Figure 2—source data 1
This spreadsheet contains the quantification and statistical analysis for total ubiquitin levels (Figure 2B) and for ubiquitin degradation in a cycloheximide chase experiment (Figure 2D–E).
- https://doi.org/10.7554/eLife.29176.011
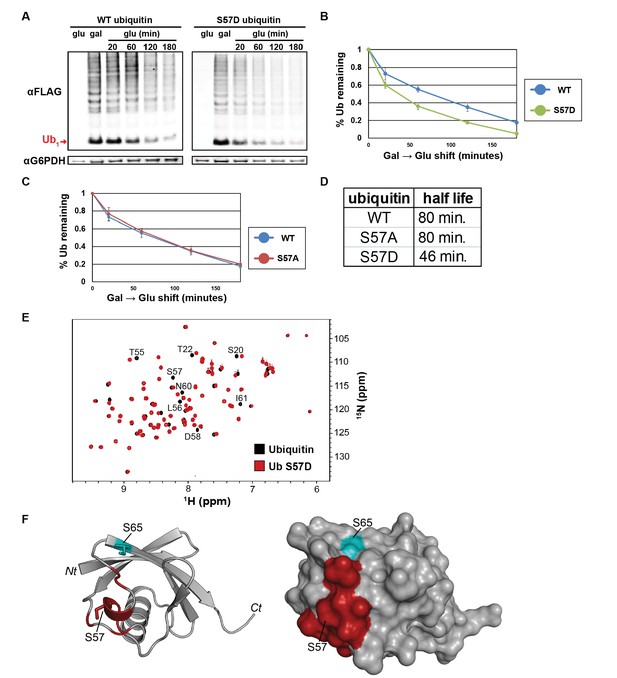
The Ser57 position of ubiquitin is a critical determinant of ubiquitin metabolism.
(A) SDS-PAGE immunoblot analysis of lysates from cells expressing FLAG-tagged ubiquitin from the GAL10 galactose-inducible promoter. Yeast strains were grown to mid-log phase in glucose media, induced to express FLAG-ubiquitin by shifting to galactose-containing media overnight, and then shifted back to glucose-containing media to repress further transcription of FLAG-ubiquitin. The top panel shows α–FLAG immunoblots and the bottom panel shows immunoblots of G6PDH, a loading control. Mono-ubiquitin is indicated by the red arrow. (B and C) Quantitation of ubiquitin degradation for wildtype, S57D (B) and S57A (C) ubiquitin was averaged (n = 4) with error bars indicating standard deviation. (D) Ubiquitin half-life was estimated based on trendline regression analysis. (E) Overlay of 15N-1H HSQC spectra generated for wild-type ubiquitin (black) and Ser57Asp phosphomimetic ubiquitin (red). Residues that are significantly perturbed in the phosphomimetic are labeled. (F) Residues perturbed in the Ser57Asp phosphomimetic (from Figure 3E) were mapped onto the structure of ubiquitin (PDB entry 1UBQ) in red. Key phosphorylation sites at Ser57 and Ser65 (cyan) are labeled.
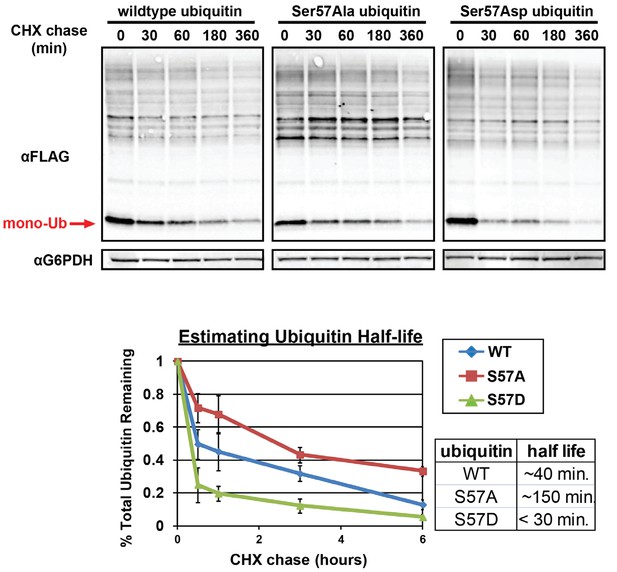
Analysis of ubiquitin half-life by SDS-PAGE immunoblot analysis of yeast lysates following a cycloheximide (CHX) chase (top panel).
Yeast strains used in these experiments express FLAG-tagged ubiquitin from the endogenous RPS31 locus. Quantitation of ubiquitin degradation (bottom panel) was averaged (n = 3) and error bars indicated standard deviation. Half-life was estimated in each case using trendline regression (table, inset bottom right).
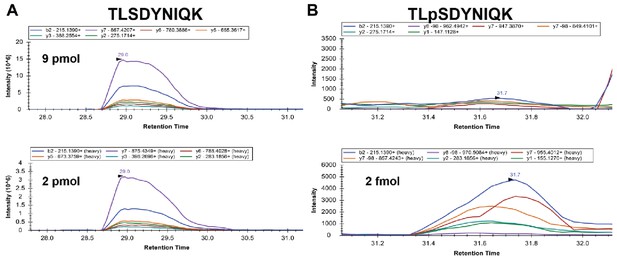
Analysis of stoichiometry of Ser57 phosphorylation of ubiquitin.
Isotopically labeled (heavy) standard peptides corresponding to unmodified (bottom left) or Ser57 phosphorylated (bottom right) ubiquitin were spiked into ubiquitin affinity purified from ppz mutant yeast cells and multiple reaction monitoring analysis was performed. The top panels show the corresponding signal from the biological samples. Inset legends indicate plots for specific transitions. Based on transition peak areas, we calculate 9 pmol of the unmodified peptide in the biological sample. The phosphopeptide in the biological sample is below our limit of quantification but occurs at significantly less than 2 fmol.
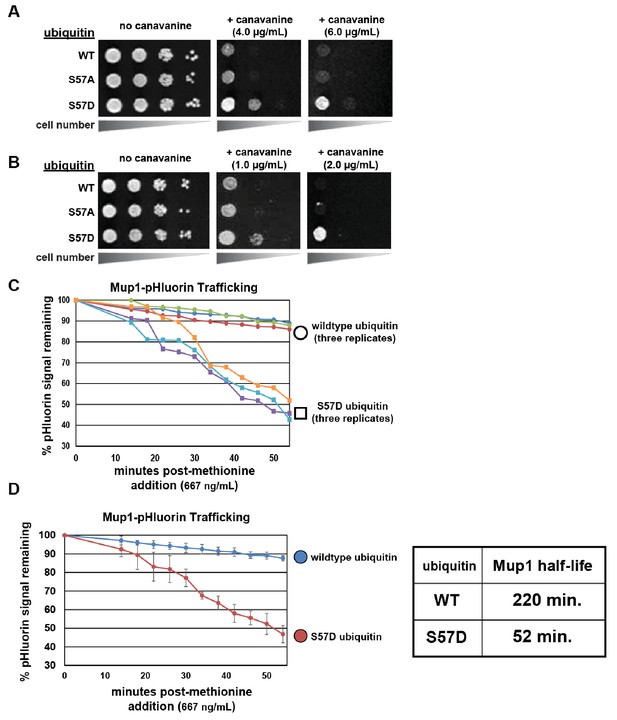
Ser57 phosphomimetic mutations in ubiquitin accelerate endocytic trafficking.
(A) Analysis of yeast growth in the presence of canavanine. In this experiment, the indicated ubiquitin variants (wildtype, Ser57Ala, or Ser57Asp) were expressed exogenously from the pRS416 plasmid under the control of the ADH1 promoter in the SEY6210 strain background. Canavanine hypersensitivity is indicative of an endocytic defect, while canavanine resistance is indicative of hyper-active endocytic trafficking. (B) Analysis of yeast growth in the presence of canavanine. Here, the indicated ubiquitin variants (wildtype, Ser57Ala, or Ser57Asp) were expressed as the sole source of ubiquitin from the native RPS31 promoter in the SUB280 strain background. (C) Flow cytometry analysis of yeast cells expressing Mup1-pHluorin grown in the absence of methionine and then stimulated by addition of a low concentration of methionine (667ng/mL). Fluorescence of pHluorin is pH-sensitive and lost during endocytic trafficking when the cargo encounters an acidic environment. Triplicate experiments are shown for cells expressing wildtype (circles) or S57D phosphomimetic (squares) ubiquitin. (D) Data generated in (C) were averaged (n = 3, error bars indicate standard deviation) and linear regression was used to generate trend lines and estimate the half-life of Mup1-pHluorin in the context of wildtype or Ser57Asp ubiquitin, as indicated in the table (right).

Hyper-active endocytic trafficking in the presence of Ser57Asp phosphomimetic ubiquitin requires conjugation.
In this experiment, the indicated ubiquitin variants (wildtype, Ser57Asp, and Ser57Asp combined with mutations that prevent ubiquitin conjugation) were expressed exogenously from the pRS416 plasmid under the control of the ADH1 promoter in the SEY6210 strain background. Canavanine hypersensitivity is indicative of an endocytic defect, while canavanine resistance is indicative of hyper-active endocytic trafficking.
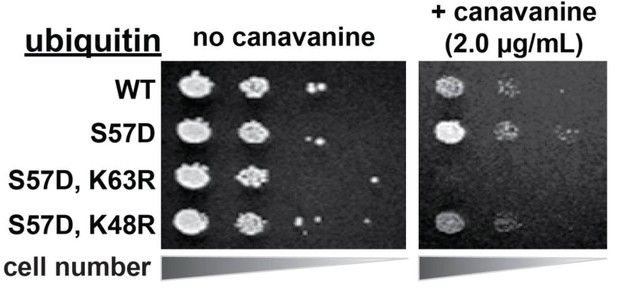
Hyper-active endocytic trafficking in the presence of Ser57Asp phosphomimetic ubiquitin requires K63-linked poly-ubiquitination.
In this experiment, the indicated ubiquitin variants (wildtype, Ser57Asp, and Ser57Asp in combination with mutations that restrict the formation of specific polyubiquitin linkage types) were expressed exogenously from the pRS416 plasmid under the control of the ADH1 promoter in the SEY6210 strain background. Canavanine hypersensitivity is indicative of an endocytic defect, while canavanine resistance is indicative of hyper-active endocytic trafficking.
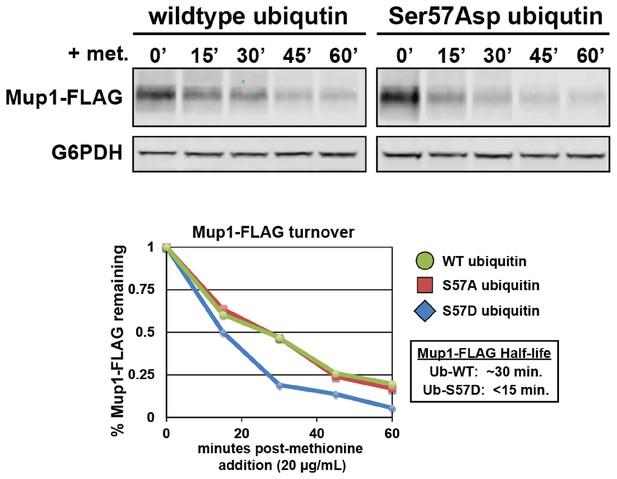
Expression of Ser57Asp phosphomimetic ubiquitin accelerates methionine-stimulated endocytic trafficking of Mup1.
Top panel shows immunoblot analysis of FLAG-tagged Mup1 following stimulation of cells by addition of methionine. Bottom panel shows the quantitation of these Mup1 turnover experiments performed in the presence of wildtype (green circles), Ser57Ala (red squares), or Ser57Asp ubiquitin (blue diamonds).
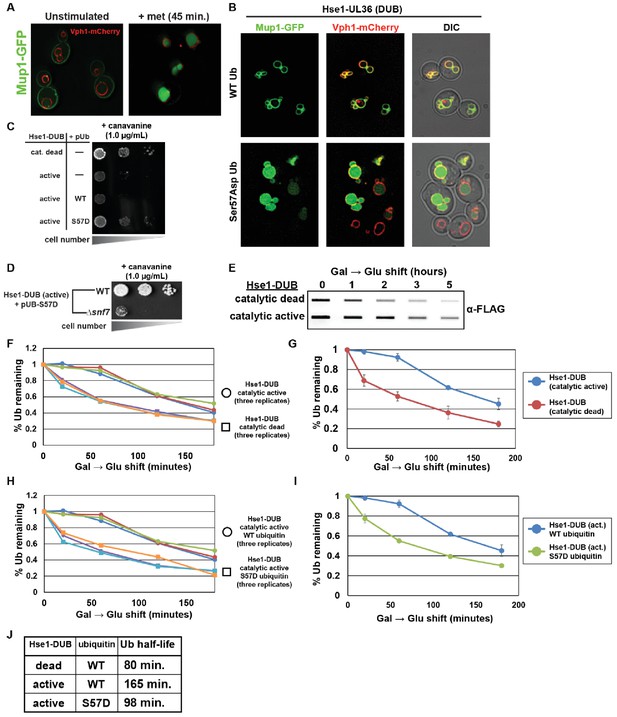
Ser57Asp phosphomimetic ubiquitin bypasses an artificial DUB checkpoint.
(A) Analysis of Mup1-GFP (green) localization in wildtype yeast cells (strain SEY6210) grown in minimal media lacking methionine (left) or stimulated with methionine for 45 min (right). Cells are stably expressing Vph1-mCherry (red), a marker of the vacuolar membrane. (B) Mup1-GFP localization in yeast cells (strain SEY6210) where the endogenous Hse1 locus has been fused to the UL36 deubiquitylating enzyme, encoding an Hse1-UL36 fusion protein. This creates an artificial DUB checkpoint at ESCRT-0. The top panel of cells are expressing wildtype ubiquitin, whereas the bottom panel of cells are expressing Ser57Asp phosphomimetic ubiquitin. As in (A), cells are stably expressing Vph1-mCherry (red), a marker of the vacuolar membrane. (C and D) Analysis of yeast growth in the presence of canavanine. (E) Slot blot analysis of FLAG-ubiquitin levels following a galactose induction/glucose repression. (F–J) Quantitation of slot blot experiments (as shown in E, n = 3) from yeast cells expressing catalytic active or catalytic dead Hse1-UL36 fusion (F and G) or catalytic active Hse1-UL36 in the presence of wildtype or Ser57Asp phosphomimetic ubiquitin (H and I). Data were averaged over multiple experiments (G and I) (n = 3, error bars indicate standard deviation) and trend line regression was used to calculate ubiquitin half life (J).

Ser57Asp phosphomimetic ubiquitin does not alter the rate of Rsp5-mediated conjugation in vitro.
In this experiment, recombinant yeast proteins GST-Uba1 (E1), 6xHIS-Ubc1 (E2), 6xHIS-Rsp5 (E3), Art1-FLAG (substrate), and ubiquitin or Ser57Asp phosphomimetic ubiquitin were incubated at room temperature in the presence of ATP. Each conjugation time course was analyzed by α-FLAG immunoblotting and quantified over three independent experiments (n = 3).
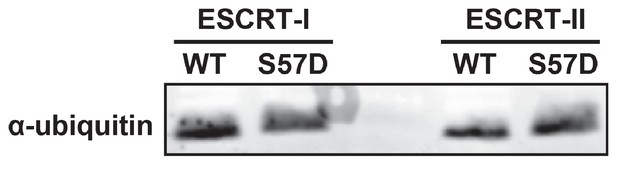
Ser57Asp phosphomimetic mutation does not alter binding of ubiquitin to ESCRT-I or ESCRT-II in vitro.
In this experiment, recombinant ESCRT-I and ESCRT-II complexes were used as bait in the capture of wild-type or Ser57Asp phosphomimetic ubiquitin. Captured samples were analyzed by SDS-PAGE followed by immunoblotting for total ubiquitin.
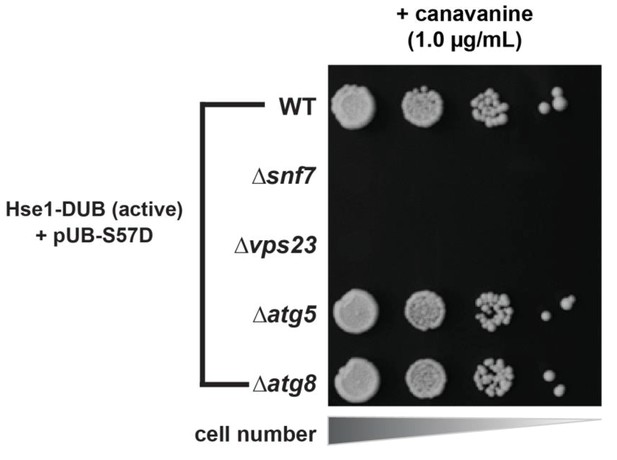
DUB bypass by Ser57Asp phosphomimetic ubiquitin requires an in-tact ESCRT pathway while the autophagy pathway is dispensible.
Yeast strains (SEY6210 background) expressing Hse1-UL36 fusions and Ser57Asp phosphomimetic ubiquitin were analyzed for growth in the presence of canavanine in order to determine the genetic requirements for the canavanine resistance phenotype.
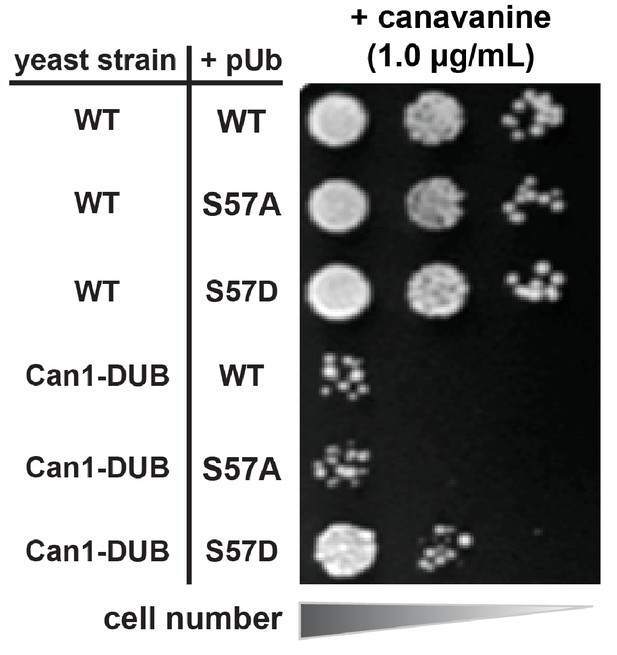
Bypass of a Can1-DUB fusion by Ser57Asp phosphomimetic ubiquitin.
Wild-type yeast strains (SUB280 background) or an isogenic strain encoding an endogenous Can1-UL36 DUB fusion were analyzed for canavanine sensitivity/resistance in the presence of wild-type, Ser57Ala, or Ser57Asp phosphomimetic ubiquitin.
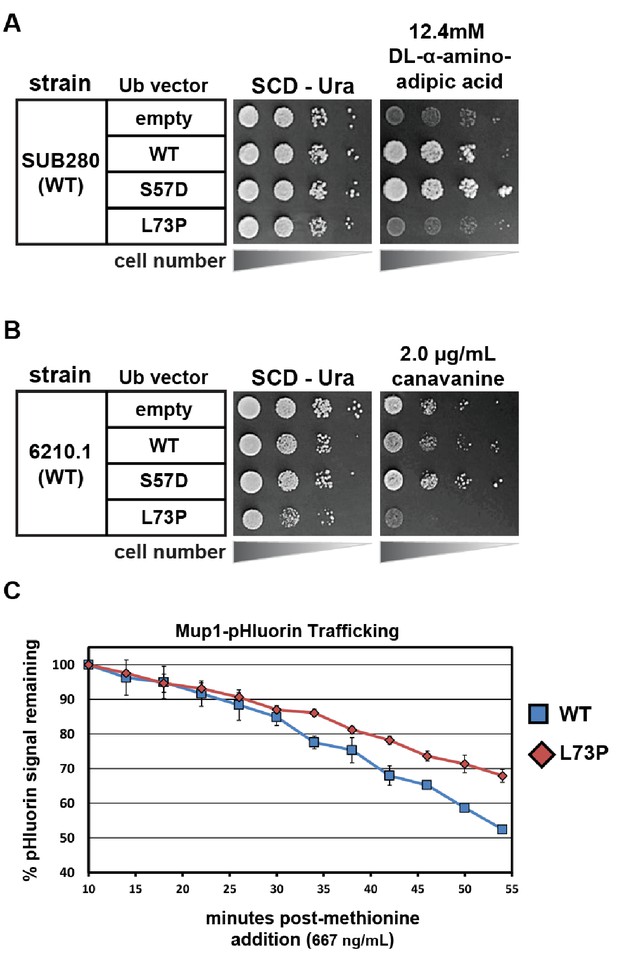
Phenotypic analysis of yeast cells expressing Leu73Pro ubiquitin.
(A) SUB280 yeast cells (expressing ubiquitin from a single pLYS source plasmid) were transformed with a second ubiquitin-expressing plasmid (pURA) and grown in the presence of DL-α-aminoadipic acid to counterselect against the pLYS plasmid (‘plasmid shuffle’). (B) Wildtype yeast cells (SEY6210.1 background) transformed with either empty vector or plasmids expressing ubiquitin (wildtype, S57A or L73P) from the ADH1 promoter were plated on minimal media with or without canavanine. (C) Wildtype yeast cells expressing pHluorin-tagged Mup1 (SEY6210.1 background) transformed with either empty vector or plasmids expressing ubiquitin (wildtype, S57A or L73P) from the ADH1 promoter were monitored using flow cytometry measurements of pHluorin signal in a time course following methionine stimulation.
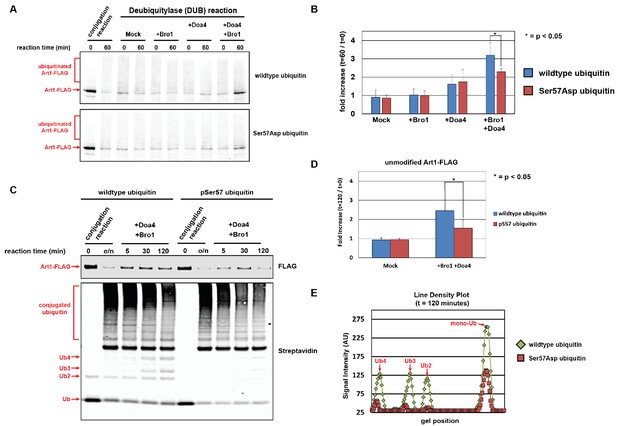
Ubiquitin phosphorylated at Ser57 is less susceptible to cleavage by Doa4.
(A) Analysis of in vitro reconstituted Doa4 activity toward a substrate (Art1-3xFLAG, 50 nM) that has been conjugated to either wild-type or Ser57Asp phosphomimetic ubiquitin (see Figure 5—figure supplement 1). Deubiquitylation reactions were performed for 60 min in the presence of 40 nM 6xHIS-Doa4 (full length) and 3.2 µM Bro1 (C-terminal fragment with amino acids 388–844), where indicated. (B) Analysis of triplicate experiments as performed in (A). Doa4 deubiquitylase activity was measured as a function of the increase in intensity of the unmodified Art1 band relative to the start of the reaction. * indicates a statistically significant difference (p<0.05). (C) Analysis of in vitro reconstituted Doa4 activity toward a substrate (Art1-3xFLAG, 50 nM) that has been conjugated to either wild-type or Ser57 phosphorylated synthetic ubiquitin (Ubiquigent, Dundee, Scotland). Deubiquitylation reactions were performed in the presence of 40 nM 6xHIS-Doa4 (full length) and 3.2 µM Bro1 (C-terminal fragment with amino acids 388–844), where indicated. (D) Analysis of triplicate experiments as performed in (C). Doa4 deubiquitylase activity was measured as a function of the increase in intensity of the unmodified Art1 band relative to the start of the reaction. (E) Line density plots for the t = 120 min time point (C) were generated using ImageJ. * indicates a statistically significant difference (p<0.05).
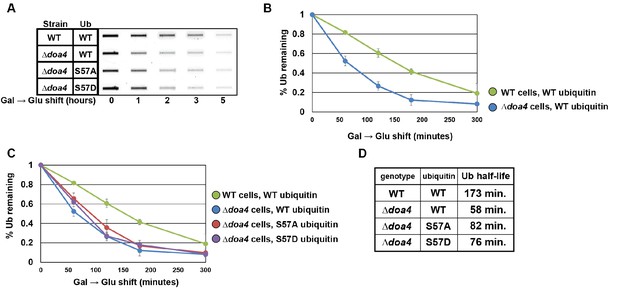
Doa4 is a key determinant of cellular ubiquitin metabolism.
(A) Slot blot analysis of FLAG-ubiquitin levels following a galactose induction/glucose repression. (B and C) Data from slot blots (as shown in A) were quantified in ImageJ and averaged (n = 4) to generate the plots of ubiquitin degradation during the glucose repression time course. (D) Data from experiments in (B) and (C) were used to calculate ubiquitin half-life using trendline regression analysis.
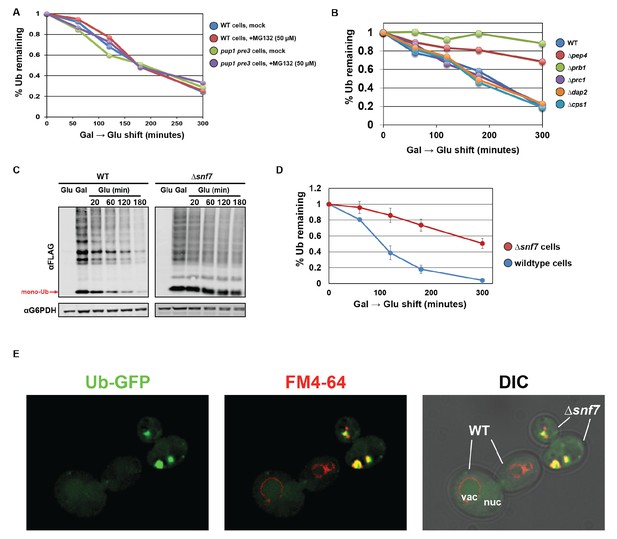
Ubiquitin is degraded in the vacuole via the ESCRT pathway.
(A) Analysis of ubiquitin half-life following galactose induction/glucose repression in wild-type or pup1pre3 mutant yeast cells (strain background WCG4A) that have been mock-treated or treated with 50 µM MG-132, a proteasome inhibitor. (B) Analysis of ubiquitin half-life following galactose induction/glucose repression in wild-type or vacuole protease mutant yeast cells (strain background BY4742). (C) SDS-PAGE and immunoblot analysis of lysates collected from yeast cells following a galactose induction/glucose repression experiment to measure ubiquitin half-life. (D) Quantitation of slot blot analysis of FLAG-ubiquitin levels following a galactose induction/glucose repression (n = 4). Error bars indicate standard deviation. (E) Fluorescence microscopy imaging of GFP-tagged ubiquitin (green) in cells where FM4-64 (red) has been chased to label the vacuole membrane. Wild-type and Δsnf7 cells were mixed prior to imaging, with the field showing both wild-type cells and Δsnf7 cells, which are distinguished by the presence of an E-compartment.

Slot blot analysis of FLAG-ubiquitin levels following a galactose induction/glucose repression.
Results shown represent a single biological replicate. Results from triplicate experiments were quantified in ImageJ and used to generate the plots, as shown in Figure 7A, B and D.

Quantitation of slot blot analysis of FLAG-ubiquitin levels following a galactose induction/glucose repression.
Results plotted are the average values (n = 3) for each time point with error bars indicating standard deviation.

Quantitation of slot blot analysis of FLAG-ubiquitin levels following a galactose induction/glucose repression.
Results are shown are averaged (n = 4) with error bars indicating standard deviation.
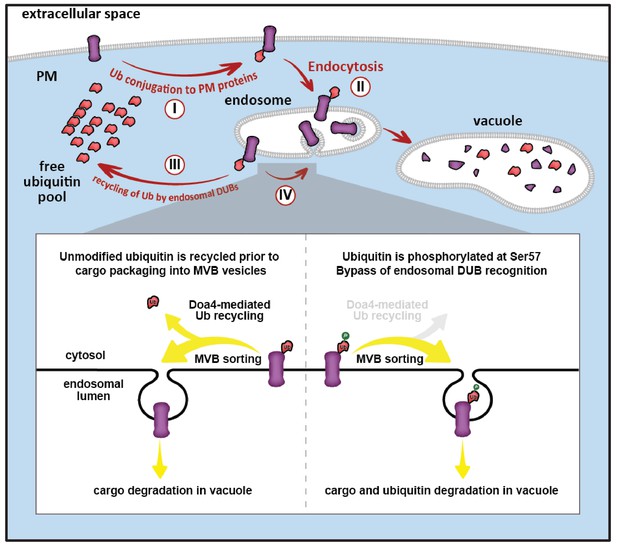
Model for ubiquitin flux through the endocytic pathway, illustrating how phosphorylation of ubiquitin at Ser57 can alter ubiquitin metabolism by limiting recycling of ubiquitin and promoting its delivery to the vacuole for degradation.
(I) Plasma membrane proteins can be conjugated to ubiquitin, resulting in sorting and internalization by endocytosis (II). Ubiquitinated cargos on the limiting membrane of the endosome can be deubiquitylated (III) resulting in recycling of both cargo and ubiquitin, or they can be captured and sorted by ESCRT machinery into vesicles that bud into the lumen of the endosome. (IV) In the final stages of the ESCRT pathway, ubiquitin can be recycled prior packaging of cargo into vesicles by Doa4, but this mode of ubiquitin recycling is abrogated by phosphorylation at the Ser57 position of ubiquitin (inset).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain background (S. cerevisiae) | SEY6210.1 | S. Emr Lab;Robinson et al. (1988) | WT: Mat a leu2-3,112 ura4-52 his3-∆200 trp1-∆901 lys2-801 suc2-∆9 | |
| Strain background (S. cerevisiae) | SUB280 | D. Finley Lab; Finley et al. (1994) | MATa, lys2-801, leu2-3, 112, ura3-52, his3-Δ200, trp 1–1, ubi1-Δ1::TRP1, ubi2-Δ2::ura3, ubi3-Δub-2, ubi4-Δ2::LEU2 [pUB39 Ub, LYS2] [pUB100, HIS3] | |
| Strain background (S. cerevisiae) | BY4742 | Dharmacon (Lafayette, CO) | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | |
| Strain background (S. cerevisiae) | WCG4a | B. Tansey Lab; Howard et al. (2012) | MATa ura3 leu2-3,112 his3-11,15 | |
| Strain background (S. cerevisiae) | WT | this paper | SEY6210.1; MATa, TRP::HTF-ubi2, TRP::HTF-ubi3, arg4Δ::Kan | |
| Strain background (S. cerevisiae) | ∆ppz1,2 | this paper | SEY6210.1; MATa, TRP::HTF-ubi2, TRP::HTF-ubi3, arg4Δ::Kan, ppz1Δ::HIS, ppz2Δ::TRP | |
| Strain background (S. cerevisiae) | WT-Ub WT;S57A;S57D | this paper | SUB280; RPS31 (UB-WT/S57A/S57D) | |
| Strain background (S. cerevisiae) | ∆ppz1,2 | this paper | SUB280; ppz1::KAN, ppz2::NAT | |
| Strain background (S. cerevisiae) | ∆ppz1,2-Ub WT;S57A;S57D | this paper | SUB280; ppz1::KAN, ppz2::NAT, RPS31 (UB-WT/S57A/S57D) | |
| Strain background (S. cerevisiae) | WT | this paper | SEY6210.1; Mup1-pHluorin::KanMX | |
| Strain background (S. cerevisiae) | ∆ppz1,2 | this paper | SEY6210.1; Mup1-pHluorin::KanMX, ppz1Δ::HIS, ppz2Δ::TRP | |
| Strain background (S. cerevisiae) | WT-Ub WT;S57A;S57D | this paper | SEY6210.1; RPS31-HTF (UB-WT/S57A/S57D) | |
| Strain background (S. cerevisiae) | WT | this paper | SEY6210.1; Mup1::HTP-TRP | |
| Strain background (S. cerevisiae) | ∆ppz1,2 | this paper | SEY6210.1; Mup1::HTP-TRP, ppz1Δ::HIS, ppz2Δ::TRP | |
| Strain background (S. cerevisiae) | Vph1-mCherry | this paper | SEY6210.1; Vph1-mCherry | |
| Strain background (S. cerevisiae) | Hse1-DUB | this paper | SEY6210.1; Hse1-DUB (HA-UL36)::NAT | |
| Strain background (S. cerevisiae) | ∆snf7 | this paper | SEY6210.1; Hse1-DUB (HA-UL36)::NAT, snf7::His | |
| Strain background (S. cerevisiae) | ∆vps23 | this paper | SEY6210.1; Hse1-DUB (HA-UL36)::NAT, vps23::His | |
| Strain background (S. cerevisiae) | ∆atg5 | this paper | SEY6210.1; Hse1-DUB (HA-UL36)::NAT, atg5::His | |
| Strain background (S. cerevisiae) | ∆atg8 | this paper | SEY6210.1; Hse1-DUB (HA-UL36)::NAT, atg8::His | |
| Strain background (S. cerevisiae) | Can1-DUB | this paper | SEY6210.1; Can1-DUB (HA-UL36)::NAT | |
| Antibody (Mouse-Monoclonal ANTI-FLAG(R) M2 antibody) | α-Flag | Sigma (St. Louis, MO) | AB_262044 | dilution 1:1000 |
| Antibody (Mouse-MAB1510) | α-Ubiquitin | Millipore (Burlington, MA) | AB_2180556 | dilution 1:1000 |
| Antibody (Rabbit-G6PDH) | α-G6PDH | Sigma (St. Louis, MO) | AB_258454 | dilution 1:10000 |
| Antibody (Rabbit-Ubiquitin-pS57) | α-pSer57 Ub | Gift from B. Brasher | dilution 1:1000 | |
| Antibody (IRDye 680RD-Goat anti-mouse) | Licor (Lincoln, NE) | Cat.# 926–68070 | dilution 1:10000 | |
| Antibody (IRDye 800CW-Goat anti-rabbit) | Licor (Lincoln, NE) | Cat.# 926–32211 | dilution 1:10000 | |
| Chemical compound, drug (Cycloheximide) | CHX | Sigma (St. Louis, MO) | Cat. # C7698 and C1988 | |
| Chemical compound, drug (L-Canavanine) | Canavanine | Sigma (St. Louis, MO) | Cat. # C1625 | |
| Chemical compound, drug (DL-2-Aminoadipic acid) | DL-2-Aminoadipic acid | Sigma (St. Louis, MO) | Cat. # A0637 | |
| Chemical compound, drug (FM4-64) | FM4-64 | Invitrogen (Carlsbad, CA) | Cat. # T3166 | |
| Chemical compound, drug (MG132) | MG132 | Apexbio (Houston, TX) | Cat. # A2585 | |
| Expression vectors for yeast (pRS416) | Sikorski and Hieter (1989) | |||
| Expression vectors for yeast (pRS416, RPS31 (UB-WT/S57A/S57D)) | Ub-WT;S57A;S57D | this paper | ||
| Expression vectors for yeast (pRS416, pADH-RPS31 (UB-WT/S57A/S57D)) | (ADH1) Ub-WT;S57A;S57D | this paper | ||
| Expression vectors for yeast (pRS416, pTDH-RPS31 (UB-WT/S57A/S57D)) | (TDH3) Ub-WT;S57A;S57D | this paper | ||
| Expression vectors for yeast (pRS416, PPZ1-HTF) | PPZ1-Flag | this paper | ||
| Expression vectors for yeast (pRS416, PPZ1-R451L-HTF) | PPZ1-R451L-Flag | this paper | ||
| Expression vectors for yeast (pRS416, RPS31 (WT/S57A/S57D), pTEF1-HTF) | (TEF1) Ub-WT;S57A;S57D | this paper | ||
| Expression vectors for yeast (pRS416, pGAL10-HTF (UB-WT/S57A/S57D)) | (GAL10) Ub-WT;S57A;S57D | this paper | ||
| Expression vectors for yeast (pRS416, MUP1-GFP) | MUP1-GFP | this paper | ||
| Expression vectors for yeast (pRS416, ART1-HTF) | ART1-Flag | this paper | ||
| Expression vectors for yeast (pRS416, pADH-RPS31, L73P) | Ub-L73P | this paper | ||
| Expression vectors for yeast (pRS416, GFP-Ub) | GFP-Ub | this paper | ||
| Software | Adobe Illustrator CS5.1 | Version 15.1.0 | ||
| Software | ImageJ | NIH | SCR_003070 | |
| Software | Licor image studio | Licor (Lincoln, NE) | SCR_015795 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29176.032





