Alternative RNA splicing in the endothelium mediated in part by Rbfox2 regulates the arterial response to low flow
Figures
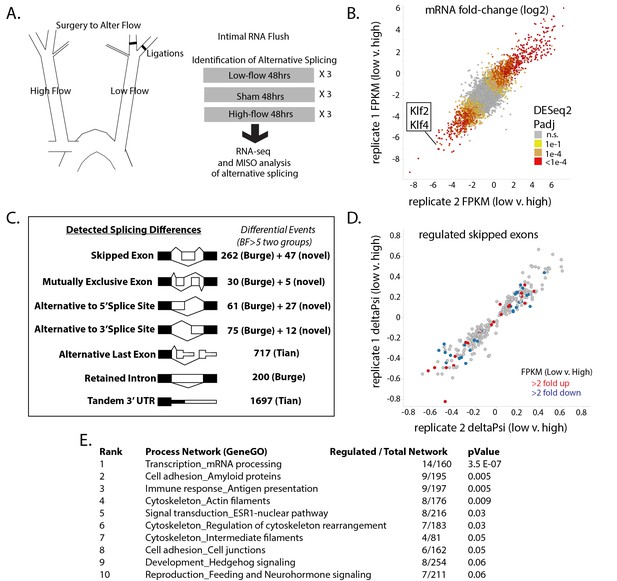
Exposure of the arterial endothelium to low and disturbed flow induces a program of alternative splicing.
RNA from the arterial endothelium was isolated 48 hr after partial carotid ligation; from the low-flow side, the high-flow side, or sham-operated vessels. (A) Outline of splicing analysis. Three pools of mRNA from each condition were isolated by polyA and sequenced. (B) Plot showing the consistency of changes in gene expression in two independent biological comparisons of low-flow versus high-flow isolations. (C) Number of RNA splicing events of each category detected by MISO analysis as significantly different in two independent biological comparisons. Events were drawn from annotated databases (Burge, Tian) or from custom annotation of mapped splice junctions (novel). (D) Plot showing the changes in skipped-exon inclusion level (deltaPsi) between low-flow and high-flow isolations. The plot also indicates changes in transcription, highlighting exons in genes with a change in FPKM of more than 2-fold up or down. (E) Processes enriched in the genes with regulated skipped exons, relative to the entire set of genes expressed in the tissue with annotated skipped exons which were not significantly regulated. Padj = Adjusted P-value, BF = Bayes Factor; FPKM = Fragments Per Kilobase of transcript per Million mapped reads; CCDS = Consensus Coding DNA Sequence.
-
Figure 1—source data 1
Contains DESeq2 output from the analysis of biological triplicates of low flow and biological triplicates of high flow, where each biological triplicate contained 4–5 carotid flushes.
DESeq2 was performed on samples following STAR alignment and sequencing counts as described in methods.
- https://doi.org/10.7554/eLife.29494.008
-
Figure 1—source data 2
Contains MISO from Tophat alignments of 80 bp reads, as described in the methods.
See GEO file GSE101826 for annotation of samples.
- https://doi.org/10.7554/eLife.29494.009
-
Figure 1—source data 3
Contains bowtie alignments to eGFP or tdTomato for the indicated samples as described in the methods.
- https://doi.org/10.7554/eLife.29494.010
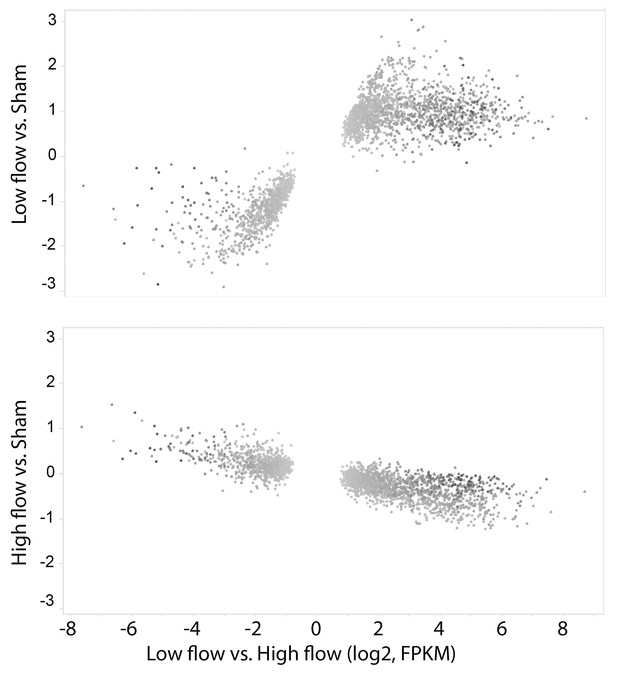
Gene expression changes between low-flow and high-flow intima are mainly due to low flow.
Alterations in transcript level for the 2535 genes with p-values<0.0001 in the comparison of low versus high flow. Most of the changes seen in the low vs high flow comparison (X-axis) were stronger in the low vs sham comparison than in the high vs sham comparison (Y-axis). All changes are shown as log2-fold.
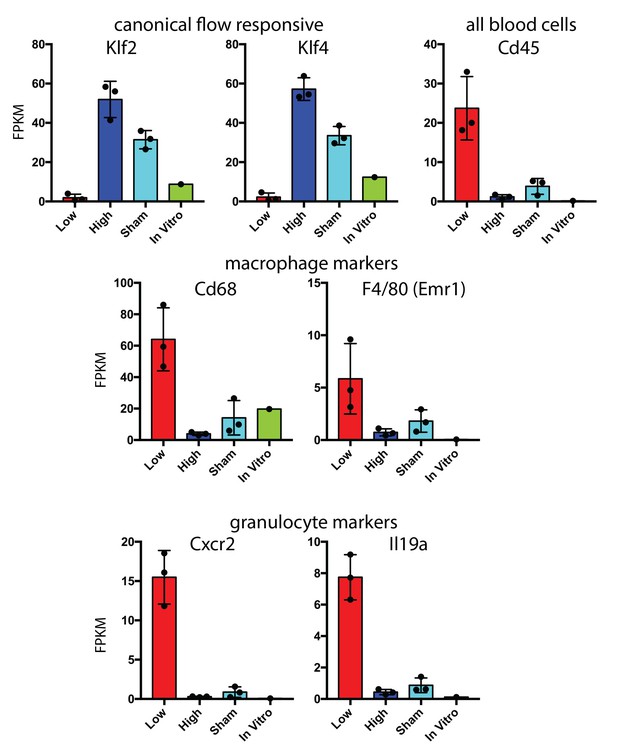
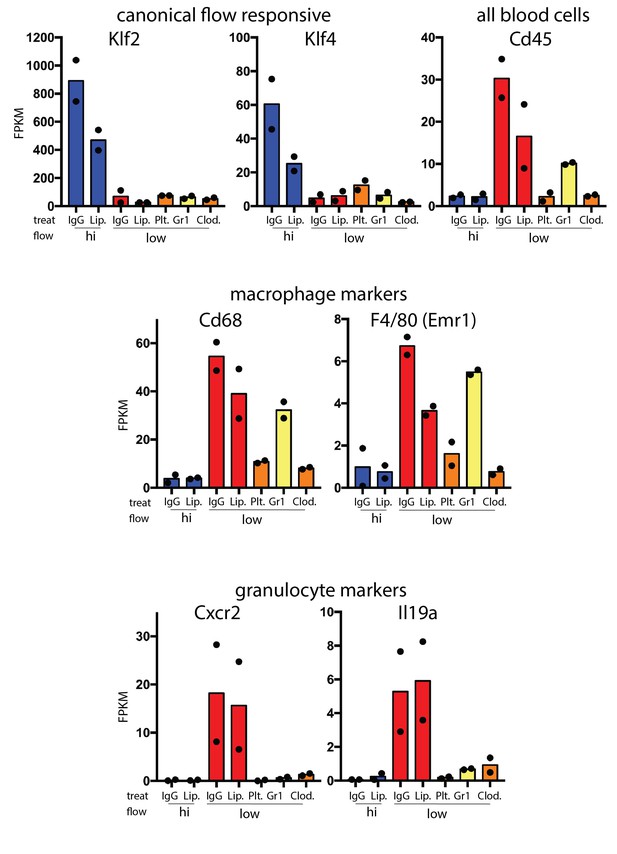
Hematopoietic cell recruitment within 48 hr of low and disturbed flow.
Comparison of RNA-seq data from carotid intima 48 hr after the induction of low and disturbed flow. Canonical flow-responsive genes (Klf2 and Klf4) and markers of recruited immune cells (CD45, Cd68, F4/80, Cxcr2 and Il19a) are shown. Each point represents pooled RNA from multiple intimal preparations. In vitro is a single isolated and sorted cell line from aortic endothelium based on eGFP expression (Cdh5(PAC)-CreERT2; mTmG).
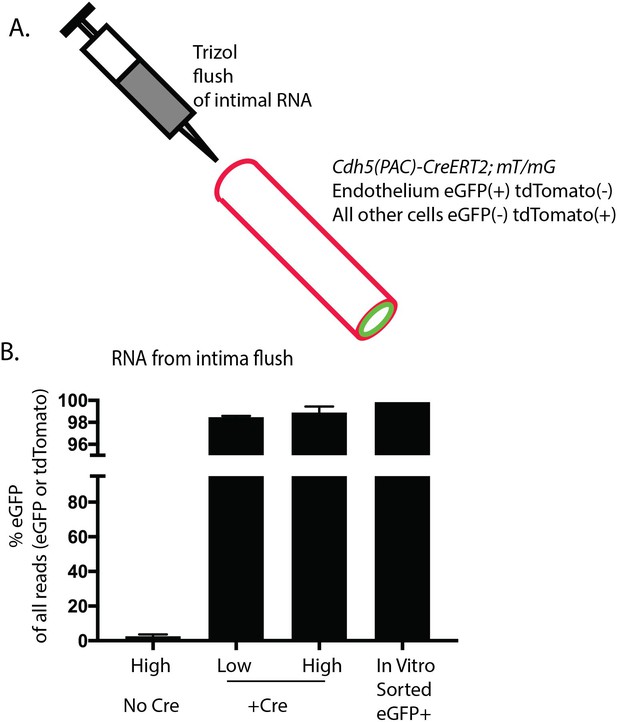
Enrichment of endothelial RNA demonstrated by lineage markers in mT/mG mice.
The mT/mG mice constitutively express tdTomato in all cells from a CMV enhancer/chicken beta-actin core promoter (pCA). Upon Cre activity, in this case driven by the endothelial-specific Cdh5(PAC)-CreERT2, the tdTomato is excised, and a downstream eGFP is expressed instead. Since tdTomato is expressed in all non-endothelial cells and eGFP is expressed in all endothelial cells, the ratio may be used as an indicator of endothelial to non-endothelial mRNA levels in the isolate. The comparison of RNA-seq reads assigned to eGFP or tdTomato from Cdh5(PAC)-CreERT2; Rbfox2 ff; mT/mG (N = 2 pools) mice or Rbfox2 ff; mT/mG mice (N = 2 pools) is shown. Cultured and FACs-purified eGFP +cells from Cdh5(PAC)-CreERT2; Rbfox2 ff; mT/mG mouse aorta is also shown (N = 1).
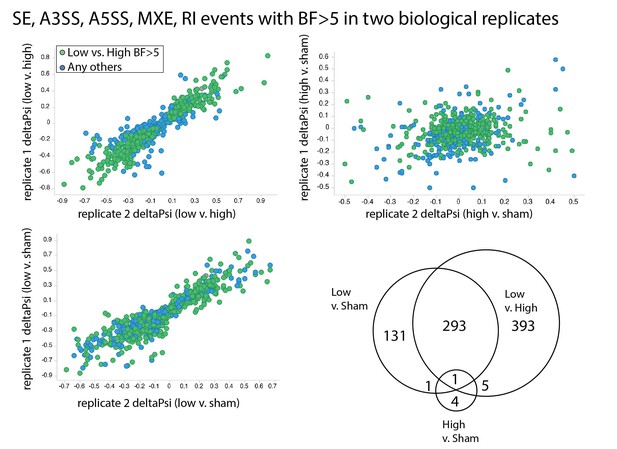
Splicing changes are primarily induced by low flow rather than high flow.
Comparison of the change in inclusion frequency by MISO for individual splicing events of the SE, MXE, A3SS, A5SS and RI classes. In green are the events presented in Figure 1C. In blue are all other events with BF >5 in two independent biological comparisons of low flow versus sham, or with BF >5 in two independent biological comparisons of high flow versus sham. Then Venn diagram shows the overlap in these events found to be significant in each of the independent comparisons.
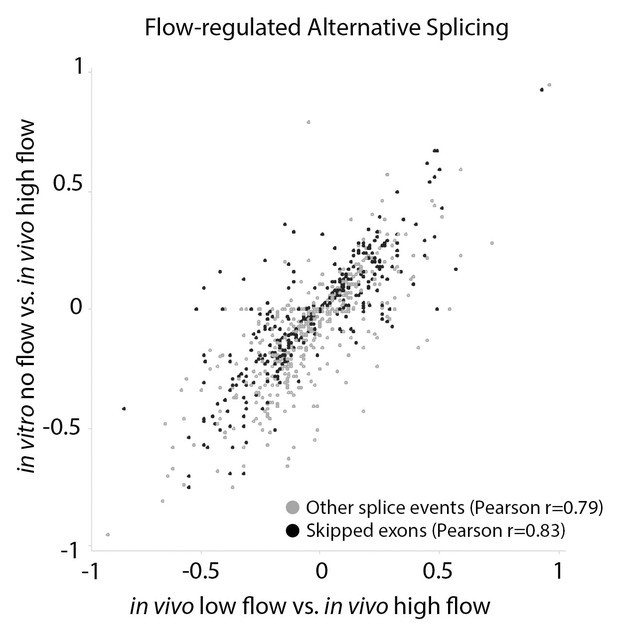
Arterial cells cultured in vitro replicate splicing changes observed under low and disturbed flow in vivo.
Comparison of the change in inclusion frequency by MISO for individual splicing events of the classes shown (by color code on bottom), relative to high-flow arterial endothelium in vivo. Other splice events include A3SS, A5SS, MXE and RI. Only flow-regulated alternative splicing events are shown. In vitro cells are primary Cdh5(PAC)-CreERT2; mT/mG cells from the aorta, isolated as described in Materials and methods, cultured in static conditions and sorted by FACs for eGFP+ before RNA isolation. Splicing events in these cells were compared to high-flow carotid artery intimal isolate. In vivo low-flow is the comparison of the low-flow intimal isolate to the high-flow intimal isolate. Pearson correlation indicates the correlation between the change in inclusion observed in low-flow activated endothelial cells in vivo versus low-flow activation of endothelial cells in vitro.
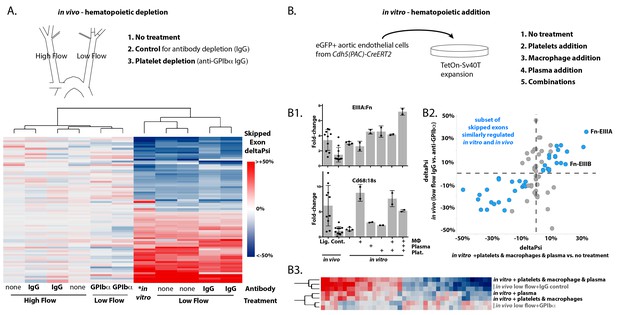
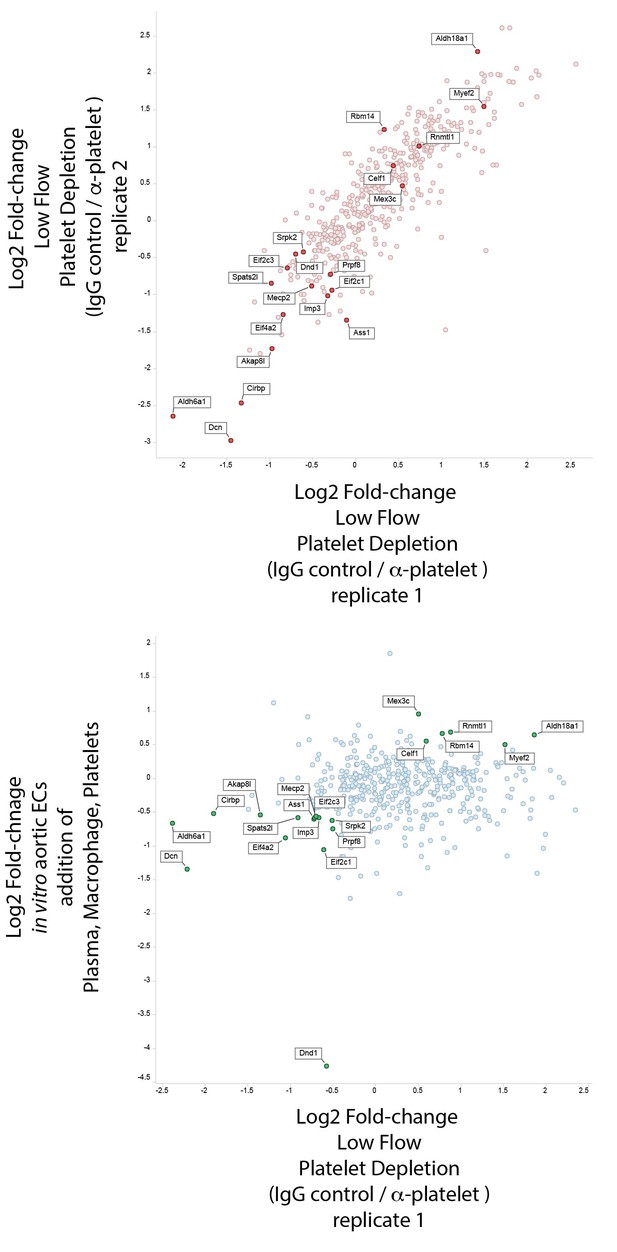
Platelets and macrophages regulate a subset of flow-regulated skipped-exon events.
(A) Effects of platelet depletion on splicing patterns in vivo (see also 2–2). Clustered heat map showing the change in skipped-exon inclusion frequency relative to high-flow contralateral arteries in biological replicates of untreated arteries (from data set created in Figure 1) or platelet-depleted (anti-GPIbα) or IgG control-treated (IgG) arteries. The data shown are for the platelet-dependent subset of all flow-regulated skipped exons (SE; 80/292). IgG and anti-GPIbα (low flow) are relative to average IgG (high flow). No antibody treatment in vitro and in vivo low flow are relative to average in vivo high flow. (B) Effects of platelet, macrophage and plasma addition to endothelial cells in vitro. (B1) Bar graphs showing in vitro changes in EIIIA inclusion frequency and Cd68 macrophage marker expression with the different treatments of conditionally immortalized aortic endothelial cells, relative to in vivo low-flow and high-flow samples. (B2) Plot showing the in vitro regulation of platelet-regulated skipped-exon events (40/80), in isolated and conditionally immortalized aortic endothelial cells. (B3) Clustered heat map, showing the change in skipped exon inclusion frequency in in vivo biological replicates of low-flow arteries with or without platelet depletion (in vivo low-flow+ IgG control, +GPIba) or conditionally immortalized aortic endothelial cells with the addition of the indicated cells or 10% plasma.
-
Figure 2—source data 1
Contains MISO from STAR alignments of 100 bp reads, as described in the methods.
See GEO file GSE101826 for annotation of samples.
- https://doi.org/10.7554/eLife.29494.015
-
Figure 2—source data 2
Contains ratio of EIIIA or EIIIB to total FN in the indicated samples.
- https://doi.org/10.7554/eLife.29494.016
Hematopoietic cell depletion in aortic intima 48 hr after induction of low and disturbed flow.
Comparison of RNA-seq data from carotid intima 48 hr after induction of low and disturbed flow, or in the contralateral arteries. Each point represents pooled RNA from multiple intimal preparations (N = 2 pools per condition, each a pool of 3–5 arteries). IgG is a non-specific antibody control for platelet (Plt.) and granulocyte (Gr1)-depleting antibodies. PBS liposome (Lip.) is a control for macrophage-depleting clodronate liposomes (Clod.).
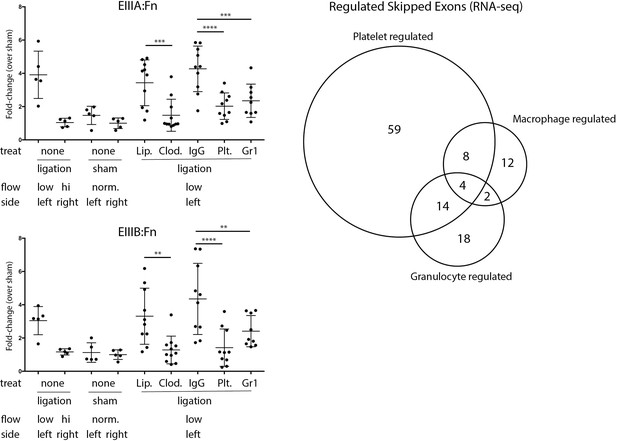
In vivo depletions of individual hematopoietic cell populations and their effects on Fn-EIIIA and –EIIIB inclusion.
Quantitative PCR results of low-flow carotid arteries isolated from mice treated with anti-Gr1 (granulocytes), anti-GPIbα (platelets), clodronate liposomes (macrophages), or controls (IgG for antibody treatments and PBS liposomes for clodronate liposomes). Change in inclusion frequency is shown as fold change relative to sham-operated control contralateral artery at 48 hr. Significance of the differences by Sidak’s multiple comparison test are shown (**p<0.01, ***p<0.001, ****p<0.0001). Venn Diagram shows the low-flow events significantly regulated in vivo by platelets, macrophages or granulocytes, as defined in methods.
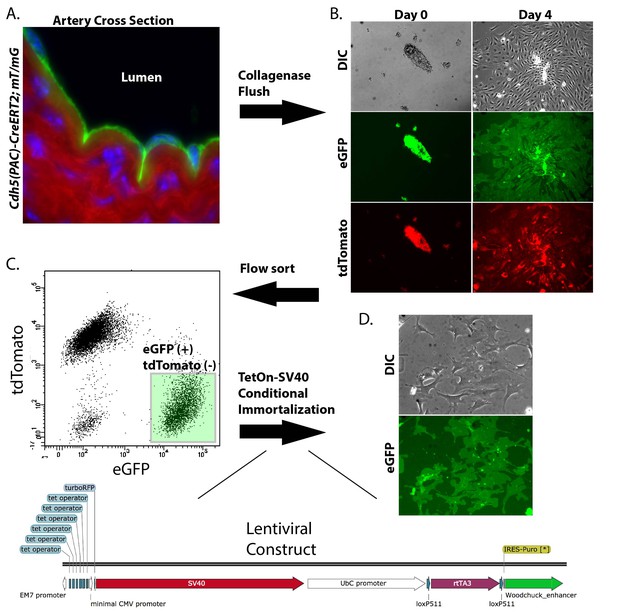
Isolation and conditional immortalization of aortic endothelial cells.
Endothelial cells were marked by induction of the mT/mG reporter in Cdh5(PAC)-CreERT2; mT/mG mice (A), isolated by collagenase flush and cultured (B), sorted on the mT/mG markers (C) and then immortalized by TetOn-SV40T (D).
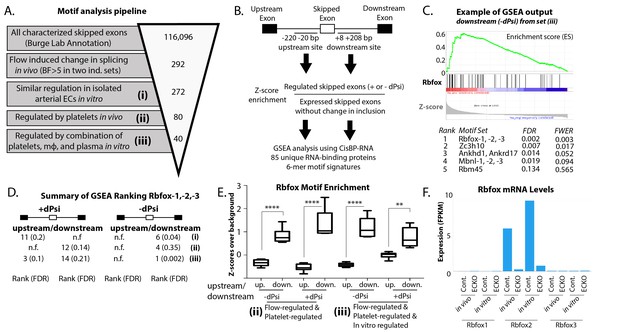
Enrichment of an Rbfox motif in the platelet/macrophage-regulated subset identified in vivo and in vitro.
(A) Motif analysis pipeline, showing the number of potentially regulated skipped-exon events, and those passing each filter. (B) Events were separated into those with increased or decreased inclusion, and then the sequences of the upstream and downstream 200 bp flanking regions were isolated, resulting in four regions of analysis. 6-mer motifs in each region were assessed for enrichment relative to exons expressed in six matched background sets of skipped exons not regulated by flow. GSEA was used to identify splicing factor ‘fingerprints’ among these enriched motifs, using the motif-binding preference defined by the CisBP-RNA database (Ray et al.). (C) Example of enrichment of Rbfox binding motif in GSEA analysis of 85-unique RNA-binding protein signatures against flanking intron sequences of flow-regulated exons. Plot shows enrichment of Rbfox family motifs among the motifs most enriched above background in the downstream flanking region of exons regulated in vivo and in vitro (set iii). (D) Enrichment of the Rbfox2 motif in each of the sets of biologically defined regulated exons (i-iii) within the upstream and downstream flanking regions of exons with either increased or decreased inclusion. (E) Plot showing the average z-score enrichment of the top 5 Rbfox in vitro defined motifs in the flanking regions of the in vivo regulated exon set (80 SE) and the in vitro regulated subset (40 SE), relative to the six matched background sets (N = 6 per bar). (F) Transcript levels of Rbfox family of proteins in the carotid artery in vivo and in isolated aortic endothelial cells in vitro, with or without the deletion of Rbfox2 by Cdh5(PAC)-CreER (EC-KO). FPKM = Fragments Per Kilobase of transcript per Million mapped reads. FDR; false-discovery rate; n.f.; not found. p<0.0001 (****), p<0.01 (**).
-
Figure 3—source data 1
Contains the pared down list of flow-regulated events and their annotations (e.g. platelet regulated).
See GEO file GSE101826 for annotation of samples.
- https://doi.org/10.7554/eLife.29494.021
-
Figure 3—source data 2
Contains the list of intervals used for motif enrichment around the indicated regulated exons for the indicated sets (e.g. increased inclusion in flow regulated set).
- https://doi.org/10.7554/eLife.29494.022
-
Figure 3—source data 3
Contains a list of expressions across samples for all known RNA binding proteins.
- https://doi.org/10.7554/eLife.29494.023
-
Figure 3—source data 4
Contains the list of all motifs, their enrichment adjacent to regulated exons, their conservation score, and the z-score enrichment of the motif from the CisBP-RNA database.
- https://doi.org/10.7554/eLife.29494.024
No obvious candidate splice factors from differential expression under altered flow conditions.
Differential expression of RNA-binding proteins under conditions with altered splicing. Factors highlighted had consistent differences of <−0.5 or >0.5 in both sets of data. Only Dcn (decorin) showed differences of <-1 in both. RNA-binding proteins with FPKM <0.5 in the in vivo data sets were filtered out.
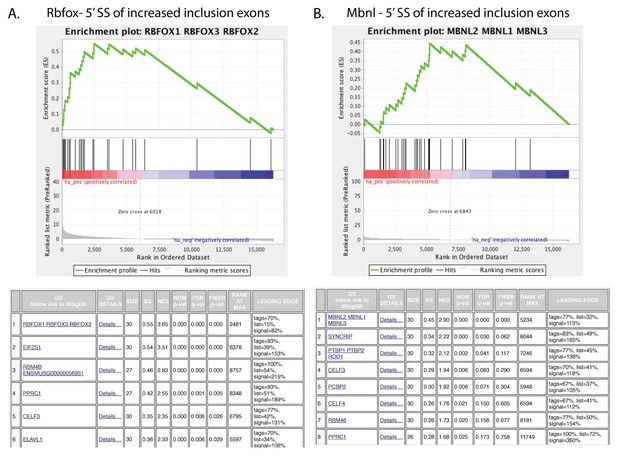
Positive control sets to confirm the motif-based identification of regulated events in vitro and in vivo by their CisBP-RNA fingerprint in GSEA.
GSEA plots showing enrichment of top CisBP-RNA motifs for the indicated splice factors near the skipped exons of the events regulated in the published gene sets (Rbfox2 shRNA in ES cells in vitro from Jiangi M et al; Mbnl1 in heart of KO mice from Wang ET et al.).
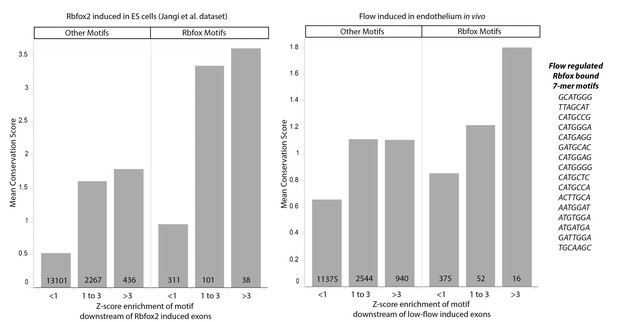
Conservation of Rbfox2 motifs enriched near flow-regulated skipped exons.
Plot showing the average conservation score (y-axis) and z-score (x-axis) for individual motifs near induced skipped exons in the published positive-control set (Rbfox2 regulated, Jangi et al. Genes and Development 2015) and the flow-induced set of skipped exon described here. Rbfox Motifs and Other Motifs are defined by Rbfox2 binding in vitro in the CisBP-RNA database (z-score >1 in CisBP-RNA). Mean conservation score on the Y-axis is derived from PhyloP vertebrate score at for each nucleotide in the given 7-mer motif adjacent to the regulated skipped exons. Z-score enrichment on the X-axis bins motifs into strongly (z-score >3), moderately (z-score 1 to 3) and weakly (z-score <1) enriched in the downstream region (5’ss) of induced skipped exons, relative to background sets of unregulated skipped exons. Number of motifs in each column are shown at the bottom of the bar. Motifs are shown for the far-right column of flow-induced (Rbfox2 motif z-score >1 in CisBP-RNA, and z-score enrichment >3 downstream of flow induced skipped exons).
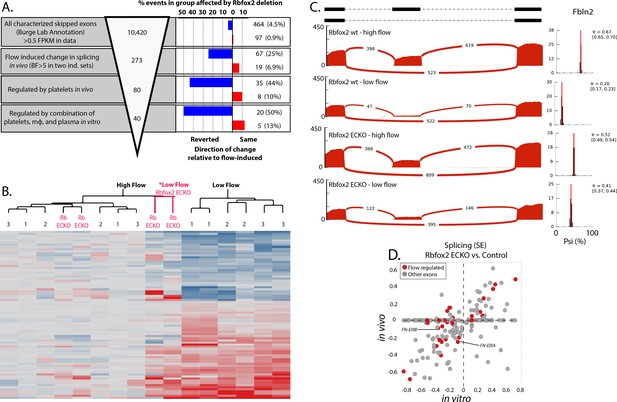
Endothelial Rbfox2 deletion affects many of the flow-regulated splicing events.
(A) Showing the percentage of each group of skipped exons regulated by endothelial deletion of Rbfox2 (BF >5) in any comparison and regulated similarly in both comparisons of Rbfox2 wt low-flow vs. Rbfox2 EC-KO low-flow – either consistent with the flow-induced change in splicing (red, ‘same’) or against the direction of the flow-induced change in splicing (blue, ‘reverted’). (B) Heat map showing the clustering of flow-regulated splicing events reverted by Rbfox2 deletion (67 of the 273 events consistently regulated between 48 hr and 7 days). 1 = C57 wild-type mice, 48 hr data set; 2 = C57 wild-type mice, IgG control 48 hr data set; 3 = Rbfox2 wt control (i.e. no Cre), 7 days data set; Rb EC-KO = Rbfox2 EC-KO 7 days data set. (C) Effect of endothelial Rbfox2 deletion on flow-mediated regulation of inclusion of Fbln2 alternative exons. (D) Plot shows the change in splicing of skipped exons (SE) following Rbfox2 deletion from carotid artery intima in vivo (under low flow) or in isolated primary aortic endothelial cells in vitro. SE from the Rbfox2 regulated set of events, in genes expressed >FPKM 1 in in vitro and in vivo sets. SE also regulated by a change in flow are highlighted.
-
Figure 4—source data 1
Contains paired list of flow-regulated skipped exon events.
Event (mm9): Skipped exon-splicing event, MM9 annotation; ENSID, RefseqID, GeneID, Rbfox2_regulated: Regulated if Rbfox2 deletion was significantly altered splicing BF >5 in any of the in vivo or in vitro comparisons; High Flow 1: Psi, 48 hr intima replicate 1 from contralateral artery pool; High Flow 2: Psi, 48 hr intima replicate 2 from contralateral artery pool; Low Flow 1: Psi, 48 hr intima replicate 1 from ligated artery pool; Low Flow 2: Psi, 48 hr intima replicate 2 from ligated artery pool; IgG High Flow 1: Psi, 48 hr intima replicate 1 from contralateral artery pool, IgG treated; IgG High Flow 2: Psi, 48 hr intima replicate 2 from contralateral artery pool, IgG treated; IgG Low Flow 1: Psi, 48 hr intima replicate 1 from ligated artery pool, IgG treated; IgG Low Flow 2: Psi, 48 hr intima replicate 2 from ligated artery pool, IgG treated; Rbfox2 wt High Flow 1: Psi, 7 days intima replicate 1 from contralateral artery pool, Rbfox2 ff; Rbfox2 wt High Flow 2: Psi, 7 days intima replicate 2 from contralateral artery pool, Rbfox2 ff; Rbfox2 wt Low Flow 1: Psi, 7 days intima replicate 1 from ligated artery pool, Rbfox2 ff Rbfox2 wt Low Flow 2: Psi, 7 days intima replicate 2 from ligated artery pool, Rbfox2 ff; Rbfox2 EC-KO High Flow 1: Psi, 7 days intima replicate 1 from contralateral artery pool, Cdh5(PAC)-CreERT2; Rbfox2 ff; Rbfox2 EC-KO High Flow 2: Psi, 7 days intima replicate 2 from contralateral artery pool, Cdh5(PAC)-CreERT2; Rbfox2 ff; Rbfox2 EC-KO Low Flow 1: Psi, 7 days intima replicate 1 from ligated artery pool, Cdh5(PAC)-CreERT2; Rbfox2 ff; Rbfox2 EC-KO Low Flow 2: Psi, 7 days intima replicate 2 from ligated artery pool, Cdh5(PAC)-CreERT2; Rbfox2 ff; Rbfox2 wt in vitro: Psi, cultured and sorted aortic endothelial cells from Rbfox2 ff; Rbfox2 EC-KO in vitro: Psi, cultured and sorted aortic endothelial cells from Cdh5(PAC)-CreERT2; Rbfox2 ff; Platelet_regulated_50percent: If platelet depletion reverted the low flow induced change in splicing by >50%; In_vitro_factor_regulated: If addition of platelets, monocytes and plasma induced splicing changes > 50% of those induced in vivo by low flow ;Rbfox2_effect: Consistent regulation, either increased or decreased, by deletion of Rbfox2 under low flow conditions in vivo; SE_data_set: Burge v2 dataset (http://miso.readthedocs.io/en/fastmiso/annotation.html) or de novo identified events (see methods).
- https://doi.org/10.7554/eLife.29494.028
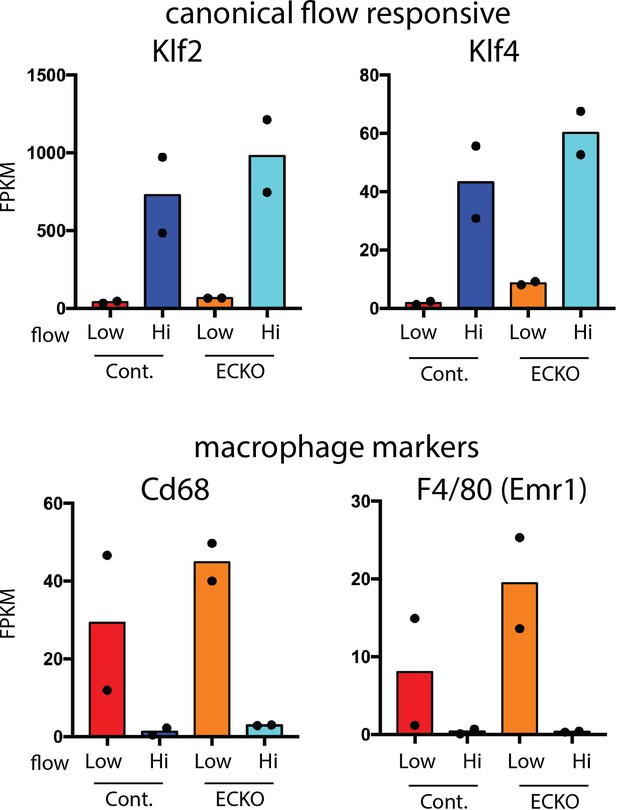
Rbfox2 deletion does not cause a reduction in markers of recruited platelets or macrophages.
Results of RNA-seq analysis of selected transcripts of intimal flush of the ligated low-flow side or the contralateral high-flow side at 7 days in Rbfox2 EC-KO mice or littermate controls.
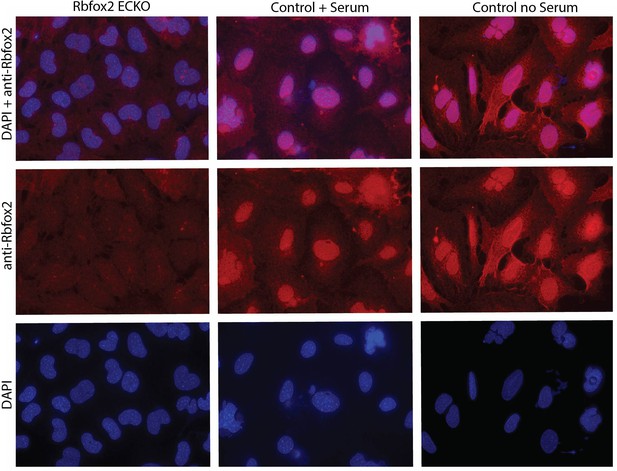
Rbfox2 localization under serum stimulation in mouse aortic endothelial cells.
Immunofluorescence staining for Rbfox2 protein in aortic endothelial cells in culture under conditions (high density + FBS) which induce EIIIA and EIIIB inclusion versus conditions which do not (high density no FBS).
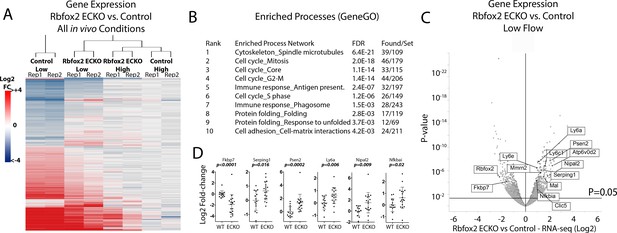
Endothelial Rbfox2 deletion suppresses low-flow transcript response in the arterial intima.
(A) Clustered heat map of the genes with adjusted p-values<0.05 shows the change in expression relative to contralateral controls at 7 days after the change in flow in the indicated genotypes (N = 641). (B) Enriched terms among the genes regulated by Rbfox2 deletion. (C) Volcano plot showing DESeq2 calculated p-values and log2 fold changes in genes expressed in the arterial intima at 7 days of arteries exposed to low and disturbed flow, with or without deletion of Rbfox2. Genes selected for qPCR in single arteries are shown. (D) Results of individual carotid artery qPCR for the genes indicated. Log2 fold-changes are relative to control, p values are from Mann-Whitney test (N = 14 control and N = 18 Rbfox2 EC-KO).
-
Figure 5—source data 1
Contains qPCR deltaCT between gene and housekeeping gene in the indicated samples, and the normalization to controls used to determine fold-change.
- https://doi.org/10.7554/eLife.29494.031
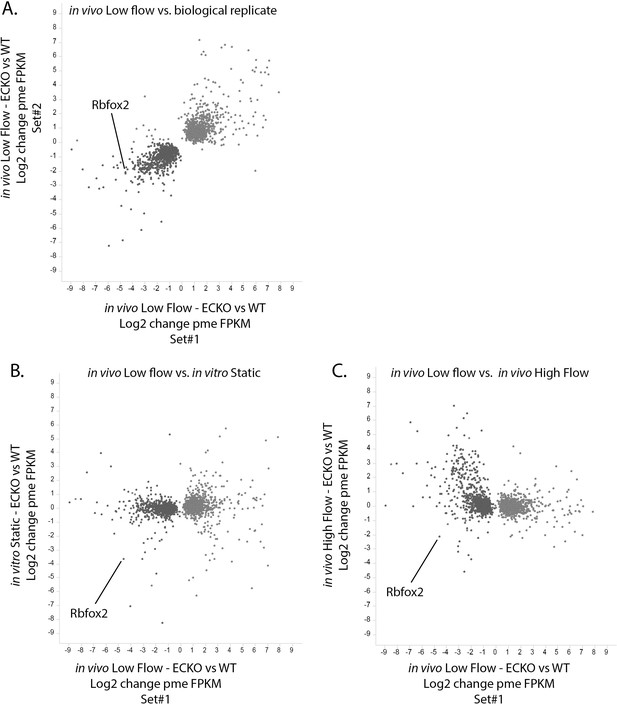
Rbfox2-dependent transcriptional response found in vivo is specific to the low-flow arterial intima.
Change in expression in the low-flow intima of Rbfox2 EC-KO mice versus littermate control mice (X-axis) plotted against the Y-axis showing (A) a biological replicate of the same comparison, (B) a comparison of isolated and cultured (7 days) aortic endothelial cells from Rbfox2 EC-KO mice versus littermate control mice, or (C) a comparison of the contralateral high- flow intima of Rbfox2 EC-KO mice versus littermate control mice. Gene expression (pme FPKM) was calculated by RSEM, and only genes with p-values of <0.05 (DESeq2) between Rbfox2 EC-KO and control under low-flow conditions are shown.
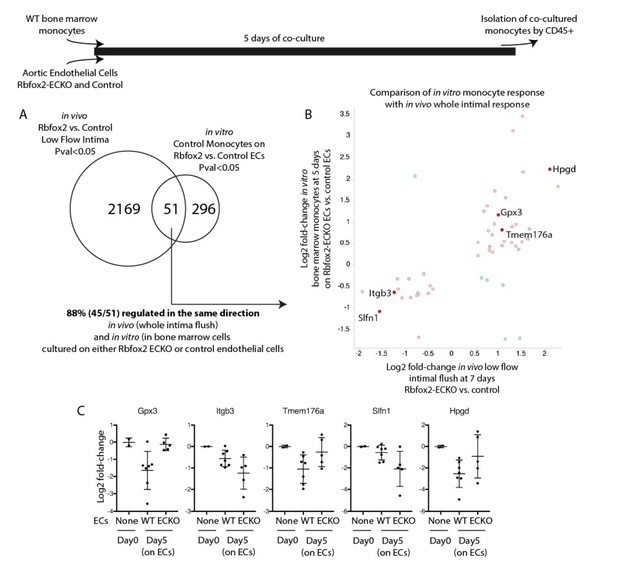
Endothelial cells deficient in Rbfox2 alter the response of co-cultured monocytes and replicate aspects of the in vivo intimal response.
Wild-type bone marrow monocytes were isolated and cultured for 5 days on Rbfox2-ECKO or control aortic endothelial cells (N=5 and N=7). Transcriptome of the initial monocytes and the co-cultured monocytes was analyzed by RNA-seq. (A) Overlap of the genes with pvalue<0.05 by DESeq2 with the genes identified by the same threshold in vivo. (B) Comparison of the fold-change observed in vitro, in co-cultured monocytes, and in vivo, in the intimal flush of low-flow carotid arteries. In red/pink are genes concordant in both, in blue are genes with opposing changes in vivo and in vitro. (C) Log2 fold-change (relative to the input monocytes at Day0) in monocytes cultured on biological replicates of Rbfox2-ECO or WT aortic endothelial cells.
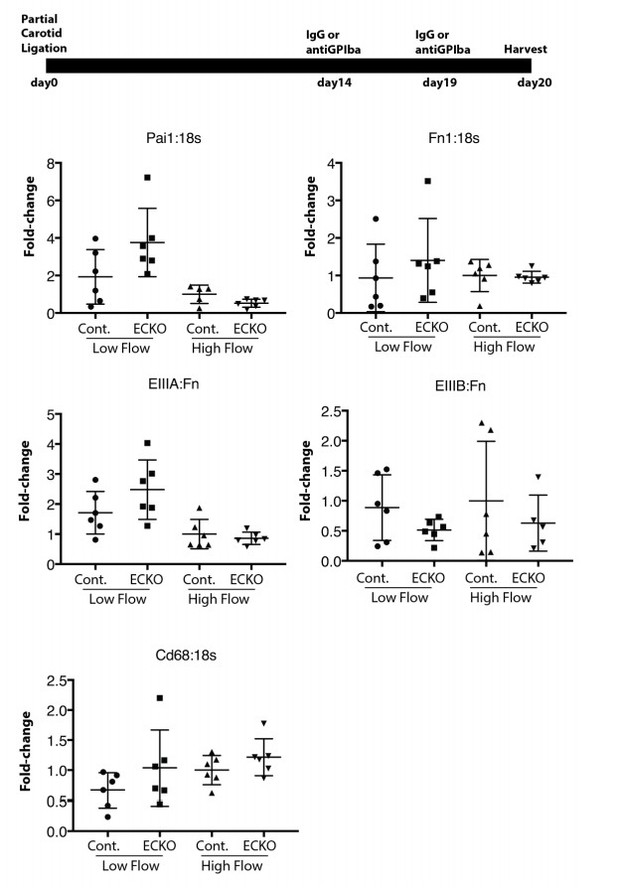
Late platelet depletion in chronic low flow injury is insufficient to reverse EIIIA splicing and PAI1 expression.
Wild-type mice were subjected to partial carotid ligation, and two weeks later platelets were depleted or not. Six days after depletion, intimal RNA flushes were obtained, and levels of gene expression assessed by qPCR.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers |
|---|---|---|---|
| antibody | anti-GP1bα | Emfret | RRID:AB_2721041 |
| antibody | anti-Gr1 | Biolegend | RRID:AB_467731 |
| antibody | anti-Rbfox2 | Bethyl Labs | RRID:AB_609476 |
| commercial assay or kit | SMARTer Universal Low Input RNA Kit | Clontech | |
| commercial assay or kit | RNAeasy microcolumns | Qiagen | |
| software, algorithm | RSEM v.1.2.15 | GitHub | RRID:SCR_013027 |
| software, algorithm | MISO v.0.4.9 | GitHub | RRID:SCR_003124 |
| software, algorithm | STAR 2.5.1b | GitHub | RRID:SCR_015899 |
| software, algorithm | Tophat 2.0.6 | GitHub | RRID:SCR_013035 |
| software, algorithm | Bowtie2 v. 2.0.5 | GitHub | RRID:SCR_005476 |
| software, algorithm | GSEA | http://www.gsea-msigdb.org/gsea/index.jsp | RRID:SCR_003199 |
| strain, strain background (M. musculus) | Rbfox2lox/lox | Jackson Lab | RRID:IMSR_JAX:014090 |
| strain, strain background (M. musculus) | Rosa26-mTmG | Jackson Lab | RRID:IMSR_JAX:007676 |
| strain, strain background (M. musculus) | Cdh5(PAC)-CreERT2 | Jackson Lab | RRID:IMSR_TAC:13073 |
| strain, strain background (M. musculus) | C57BL/6J | Jackson Lab | RRID:IMSR_JAX:000664 |
Additional files
-
Supplementary file 1
Usage of Source data.
- https://doi.org/10.7554/eLife.29494.032
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29494.033





