A gonad-expressed opsin mediates light-induced spawning in the jellyfish Clytia
Figures
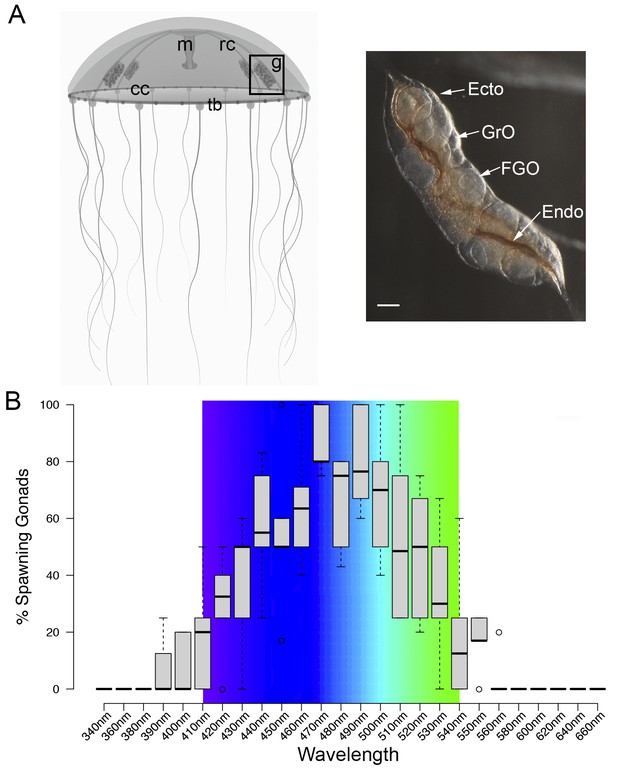
Spectral characterisation of spawning in Clytia ovaries.
(A) Schematic of a Clytia hemisphaerica female jellyfish: The four gonads (g) lie on the radial canals (rc) that connect the manubrium (M) to the circular canal (cc) running around the bell rim, adjacent to the tentacle bulbs (tb). (Inset) Photo of a Clytia ovary. FGO = fully grown oocytes; GrO = growing oocytes; Endo = gonad endoderm; Ecto = gonad ectoderm. Bar = 100 μm. (B) Box Plot showing spectral characterisation of Clytia spawning. Groups of 3–6 isolated gonads were exposed to 10 s pulses of monochromatic light (see Materials and methods). Gonads were considered to spawn if at least one oocyte underwent maturation and release. Statistics were based on percentage of gonad spawning in response to a specific wavelength obtained from 5 to 6 independent experiments. A total of 20–30 gonads were analysed per wavelength. Centre lines show the medians; box limits indicate the 25th and 75th percentiles (first and third quartiles); whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; outliers are represented by circles. Colour spectrum is shown for 410 nm – 540 nm wavelengths.
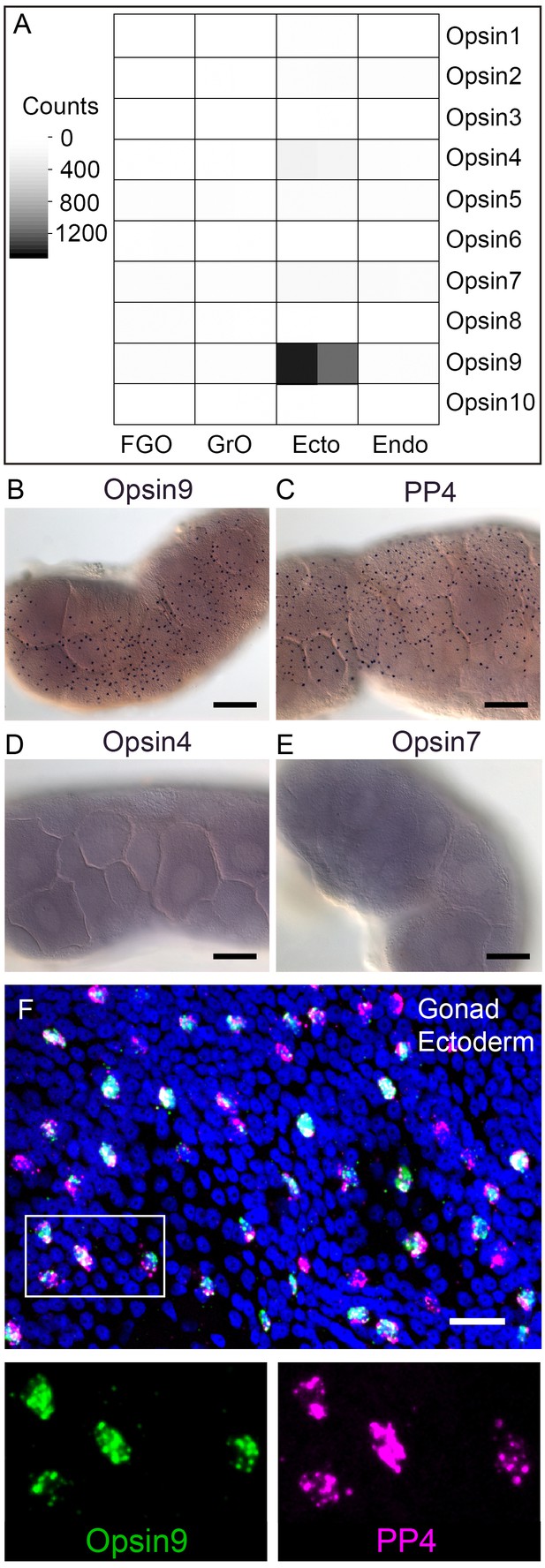
Clytia Opsin expression in gonad ectoderm cells.
(A) Heat map representing the expression of the ten opsin sequences from Clytia hemisphaerica in different gonad tissues. Illumina HiSeq 50nt reads from isolated endoderm (Endo), ectoderm (Ecto), growing (GrO) and fully-grown oocytes (FGO) from mature female ovaries were mapped against the opsin sequences. Counts were normalised per total number of reads in each sample and per sequence length (Figure 2—source data 1). (B) In situ hybridisation (ISH) detection of Opsin9 mRNA in scattered ectodermal cells of female Clytia gonads. (C) ISH of the neuropeptide precursor PP4 mRNA in Clytia female gonads, also showing a scattered pattern in the ectoderm. (D-E) ISH of Opsin4 and Opsin7 mRNAs, respectively, in Clytia ovaries, showing no detectable localised expression of these two opsin genes. (F) Double fluorescent ISH showing co-expression of Opsin9 (green) and PP4 (magenta) mRNAs in gonad ectoderm cells; nuclei (Hoechst) in blue. Single channels are shown for the outlined zone in the top image. Of n = 594 randomly chosen cells expressing either gene counted in 10 different gonads, over 86% co-expressed Opsin9 and PP4 mRNAs. Controls with single probes were performed to validate a correct fluorescence inactivation and ensure that the two channels did not cross-over (not shown). Scale bars in B-E = 100 μm; F = 20 μm.
-
Figure 2—source data 1
Expression data used to construct the heatmap in Figure 2A.
Illumina HiSeq 50 nt reads from isolated endoderm (Endo), ectoderm (Ecto), growing (GrO) and fully-grown oocytes (FGO) from mature female ovaries were mapped against the ten Clytia opsin sequences. Counts are normalised per total number of reads in each sample and with respect to sequence length.
- https://doi.org/10.7554/eLife.29555.006
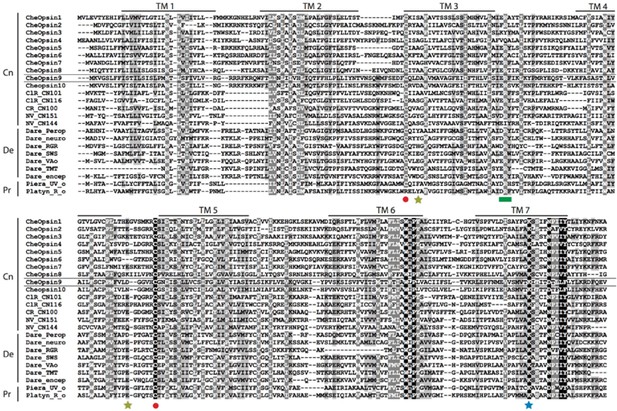
Alignment of selected opsin sequences, highlighting amino acids crucial for light detection and opsin function.
Columns of residues are highlighted by similarity group conservation (defined by GeneDoc and the BLOSUM62 matrix). Black shows 100%, dark grey shows ≥80%, and light grey shows ≥60% residues similarity in each column. Amino Acid position numbers used below correspond to the nomenclature used for bovine rhodopsin. Conserved sites are indicated for disulfide bridge formation between Cys110 and Cys187 (red circles), chromophore linkage at Lys296 (blue star), acidic counterions (which can be Glu/Asp113 and/or Glu/Asp181 – yellow stars) and G protein interaction for signal transduction (green square). Transmembrane domains are also indicated (TM1-7). Abbreviations: Cn = Cnidarians; De = Deuterostomes, Pr = Protostomes; Che = Clytia hemisphaerica ; ClR = Cladonema radiatum ; CR = Carybdea rastonii ; NV = Nematostella vectensis; Dare = Danio rerio; Piera = Pieris rapae; Platyn = Platynereis dumerelii; perop = peropsin; neuro = neuropsin; RGR = retinal G protein-coupled receptor; SWS = short wavelength sensitive; VAo = vertebrate ancient opsin; TMT = teleost multiple tissue opsin; encep = encephalopsin; UV_o = ultraviolet opsin; R_o = rhabdomeric opsin.
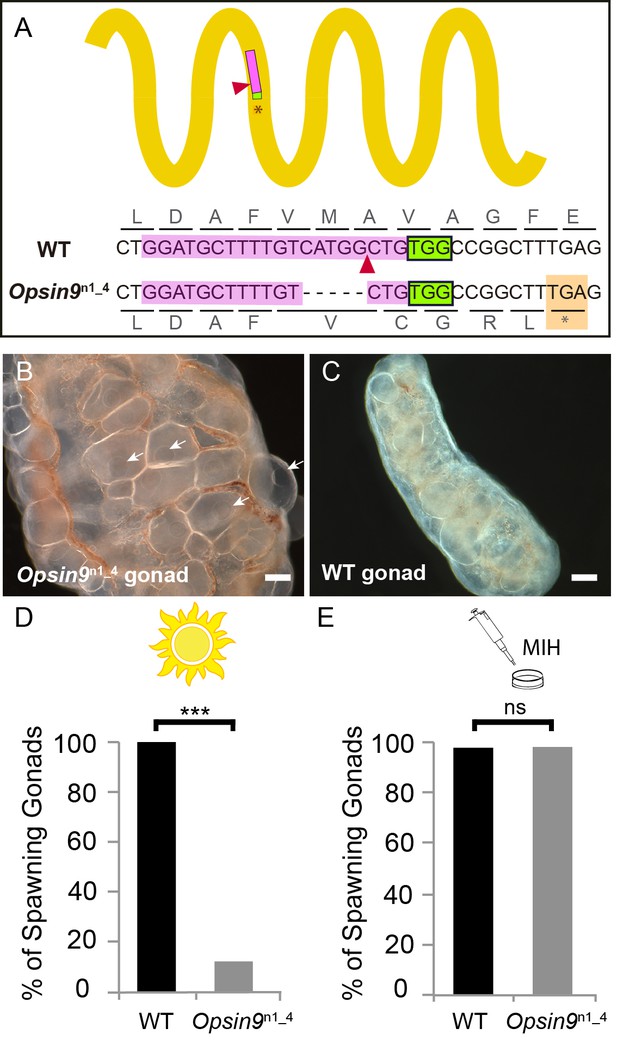
Production and phenotype of Opsin9 knockout medusae.
(A) Scheme of Opsin9 GPCR showing part of the genomic region coding for the third transmembrane domain targeted by Opsin9 n1 CRISPR sgRNA. Corresponding amino acids are shown. Pink boxes indicate the target site of the sgRNA. Green boxes are the PAM sequence (NGG). The expected cleavage site of Cas9 is indicated by red triangles. The predominant 5 bp deletion detected in the Opsin9n1_4 mutant is shown. This mutation leads to a frame-shift and an early STOP codon in Opsin9n1_4 (orange box). (B) Highly inflated gonad of an Opsin9n1_4 mutant female jellyfish showing the abnormal accumulation of large oocytes with intact GVs (arrows). (C) Wild type Clytia gonad at the same magnification. Images in B and C were both taken 8 hr after the natural light cue, accounting for the absence of fully-grown oocytes in the wild type gonad. Scale bars all 100 μm. (D) Quantification of spawning upon light stimulation of wild type (WT) and Opsin9n1_4 gonads. Percentage of spawning gonads combined from three independent experiments is shown in all cases; n = 92 for wild type and n = 154 gonads for Opsin9n1_4mutants. (E) Equivalent analysis for synthetic MIH treatment of wild type and Opsin9n1_4 gonads. Oocyte maturation and spawning were induced by synthetic MIH treatment in both cases; n = 94 gonads for wild type and n = 80 gonads for Opsin9n1_4 mutants. Fisher’s exact test showed a significant difference (at p<0.01) between spawning in wild type and mutant samples stimulated by light (D), but not by MIH (E).
-
Figure 3—source data 1
Sequences of guide RNAs and genotyping primers used in this study.
- https://doi.org/10.7554/eLife.29555.009
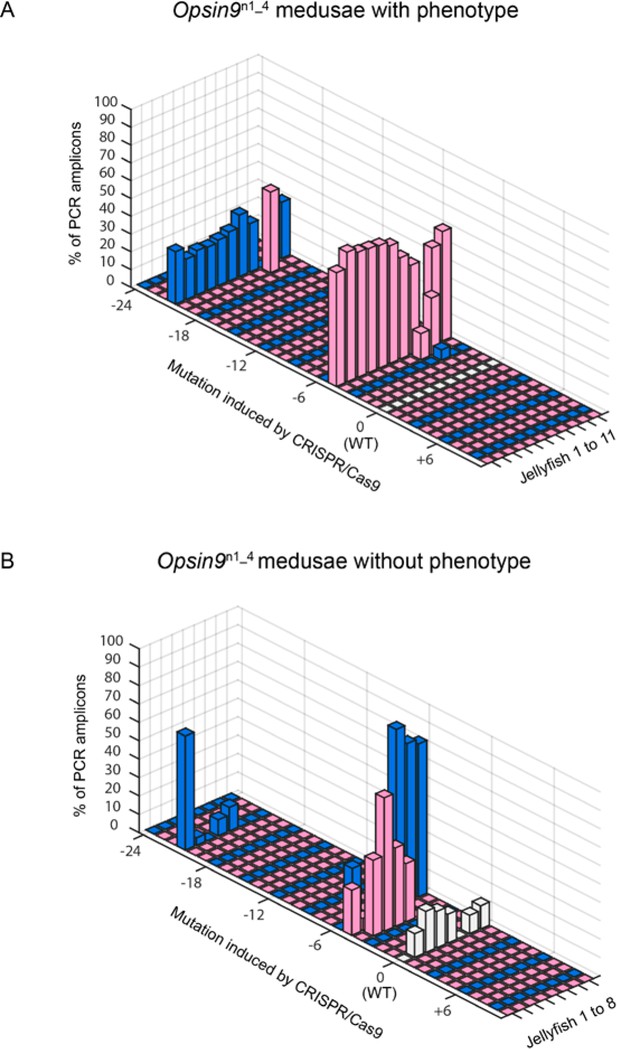
Opsin9n1_4 genotyping results.
Mutant genotyping based on PCR amplification of target site genomic areas followed by sequencing and TIDE analyses (see Materials and methods). Frame-shift mutations are shown in pink, non-frame-shift mutations in blue and wild type (WT - no mutation) in grey. (A) 3D-graph showing the mutation rate and type of 11 independent Opsin9n1_4 jellyfish with the spawning-impaired phenotype. The dominant 5 bp deletion and a long 21 bp deletion were detected in 10/11 medusae. (B) 3D-graph showing the mutation rate and type of 8 independent Opsin9n1_4 jellyfish with no phenotype. These medusae showed an increased proportion of sequences with no mutation, higher mosaicism and a lower ratio of frame-shift mutations.
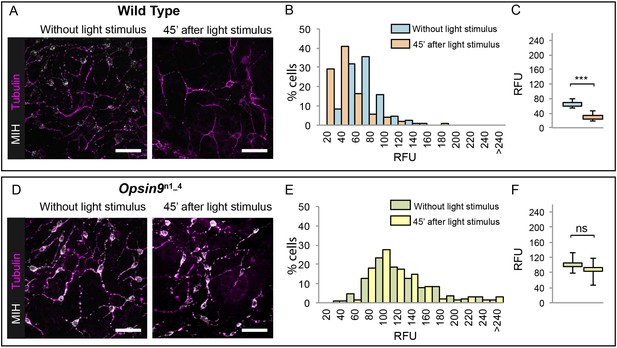
Accumulation of MIH in Opsin9 mutant gonads.
Quantification of MIH loss from gonad ectoderm after light stimulation. All images were summed from 10 confocal Z planes over 4 µm, acquired on the same day using constant settings. Immunofluorescence of fixed, methanol –treated gonads using anti-PRPamide antibodies (MIH; white) was quantified in putative MIH-Opsin cells identified by typical morphology revealed by anti-alpha tubulin (magenta) antibodies. (A) Representative images of wild type Clytia gonad showing reduction in MIH fluorescence after light stimulation. (B) Distribution of RFU (Relative Fluorescence Units) values obtained for each cell analysed in the two conditions (number of cells analysed: n = 226 and n = 282, respectively). (C) Graph showing the medians of the data in (B). Limits correspond to first and third quartiles. The Mann-Whitney U test showed a significant difference between conditions (at p<0.01). (D) Representative immunofluorescence images of Opsin9n1_4Clytia gonad MIH-secreting cells before and after light stimulation. MIH fluorescence is maintained upon light stimulation. (E) Distribution of RFU values after fluorescence quantification in Opsin9n1_4 putative MIH-Opsin cells in the two conditions (n = 183 and n = 201, respectively). (F) Graph showing the medians and quartiles of the data in (E). Mann-Whitney U test did not show a significant difference at p<0.01. Scale bars all 20 μm.
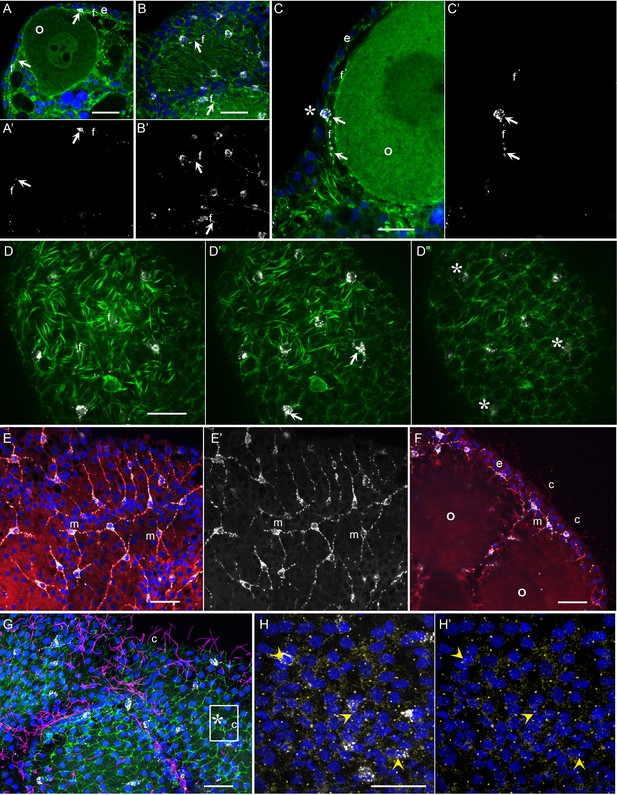
Morphology of Clytia MIH-secreting cells.
Confocal images of fluorescence stained isolated Clytia ovaries. Throughout the figure, anti-PRPamide (MIH) staining is shown in white and Hoechst staining of DNA in blue. Colours and annotations are consistent across panels. A’, B’ and E’ show PRPamide staining only , corresponding to the overlays in A, B and E. (A-D) Relationship between MIH-producing cells and the myoepithelial cells of the gonad ectoderm revealed by phalloidin staining of f-actin (green). The position of the basal layer of myofibrils is indicated by f, and examples of anti-PRPamide stained cell bodies and basal extensions are shown by white arrows. (A) Cross-section through an early growth-stage oocyte (o) and overlying ectoderm (e). (B) Glancing section through the ectodermal layer covering a largeroocyte, illustrating the characteristic phalloidin staining of basal myofibrils (f) in the centre of the image, and of the polygonal network of apical junctions in the peripheral parts of the image. (C) Higher magnification cross section of a late growth-stage oocyte and overlying ectoderm. The apical tip of anti-PRPamide stained cell lies close to the exposed surface of the ectoderm (asterisk), and the basal projections run in the myofibril layer. (D-D’) Three confocal sections taken at 1 µm intervals at progressively more superficial levels of gonad ectoderm, positioned above a large oocyte. The basal myofibril layer of the epitheliomuscular cells predominates in D and their apical junctions in D’. MIH cell bodies are centred between these two layers (D’). At the gonad surface (D”) anti-PRPamide staining is detectable at interstices between ectodermal cells (asterisks). (E-F) Anti-alpha tubulin staining (red) of methanol extracted samples highlights the microtubule bundles characteristic of the basal processes of the multipolar PRPamide-stained cells (m) and apical cilia (c) of the ectodermal cells. (E) Confocal plane at the basal ectoderm level. (F) maximum projection of 5 confocal planes (0.8 µm intervals) through oocytes and overlying ectoderm. (G) Anti- acetylated alpha-tubulin (magenta) of cilia combined with phalloidin staining (green) in a grazing superficial confocal section. A possible example of a cilium associated with the apical tip of a PRPamide-stained cell is highlighted. (H) Anti-ɣ tubulin staining (yellow) in a superficial confocal section showing apical ciliary basal bodies in each epitheliomuscular cell and in PRPamide-stained cells (arrowheads) - image overlaid in H but not H’. Scale bars all 20 μm.
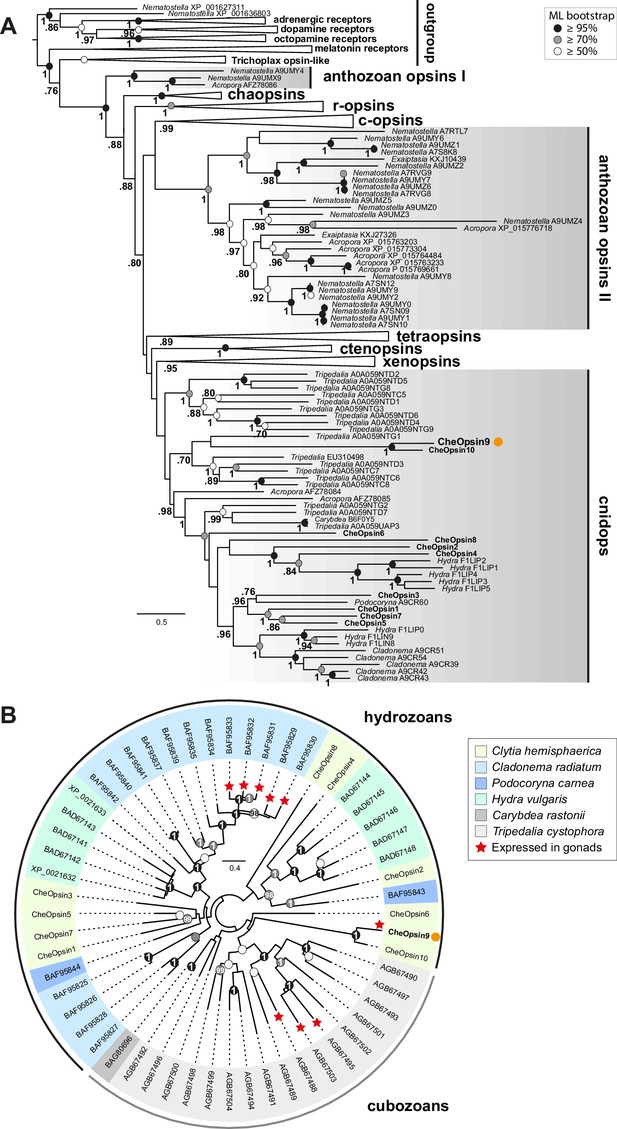
Phylogenetic position of Clytia opsins.
(A) Maximum likelihood (ML) phylogenetic analysis (RAxML, LG+Γ, Figure 6—source data 2) of opsin proteins based on the dataset of Vöcking et al. (2017) including the 10 Clytia opsins, rooted with GPCR families closely related to the opsin family. (B) Unrooted ML phylogenetic analysis of available medusozoan opsins (RAxML, LG+Γ, Figure 6—source data 3). In both trees, ML bootstrap support values (500 replicates) are shown as circles on the branch tips: black circles ≥95%, grey circles ≥70%, white circle ≥50%. Bayesian analyses (MrBayes, LG+Γ) reconstructed trees with similar topologies; Bayesian posterior probabilities greater than 0.70 (A) or 0.95 (B) are shown next to the branches (A) or on the nodes (B). The orange dot highlights CheOpsin9. NCBI or Uniprot reference numbers and Clytia opsin names are shown. In (A), c-opsins = ciliary opsins; tetraopsins = retinal G protein-coupled receptor, Neuropsin and Go-opsin as defined by Ramirez et al., 2016; r-opsins = rhabdomeric opsins; ctenopsins = ctenophore opsins; chaopsin = as defined by Ramirez et al., 2016. In (B), a red star indicates expression in the gonad. Scale bars: estimated number of substitution per site.
-
Figure 6—source data 1
Sequence alignment used for the phylogenetic analyses shown in Figure 6—figure supplement 2B, The ten Clytia opsin sequences, as well as an Opsin9/10 related sequence from unpublished transcriptome data of the hydrozoan medusa Melicertum octocostatum, were added to a published dataset (Vocking et al., 2017).
- https://doi.org/10.7554/eLife.29555.015
-
Figure 6—source data 2
Sequence alignment used for the phylogenetic analyses shown in Figure 6A and its Figure 6—figure supplement 2A, The ten Clytia opsin sequences were added to a published dataset (Vocking et al., 2017).
- https://doi.org/10.7554/eLife.29555.016
-
Figure 6—source data 3
Sequence alignment used for the phylogenetic analyses shown in Figure 6B, including the ten Clytia opsin sequences and medusozoan opsins available in GeneBank.
- https://doi.org/10.7554/eLife.29555.017
-
Figure 6—source data 4
Sequence alignment used for the phylogenetic analyses shown in Figure 6—figure supplement 1A and B, The ten Clytia opsin sequences were added to a published dataset (Feuda et al., 2014).
- https://doi.org/10.7554/eLife.29555.018
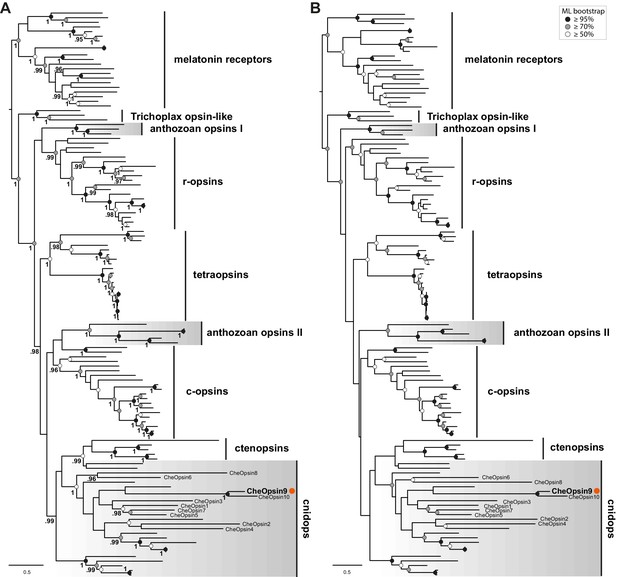
Phylogenetic analyses of the Clytia opsins based on the Feuda et al., (2014) dataset.
Maximum likelihood phylogenetic analysis of opsin proteins based on the dataset of Feuda et al. (2014) including the 10 Clytia opsins, using the (A) LG+Γ and (B) GTR+Γ models, rooted with melatonin receptors (Figure 6—source data 4). ML bootstrap support values (500 replicates) are shown as coloured circles on the branch tips: black circles ≥95%, grey circles ≥70%, white circle ≥50%. In (A), Bayesian posterior probabilities (MrBayes, LG+Γ) greater than 0.95 are shown next to the branches. Labelling as in Figure 6.
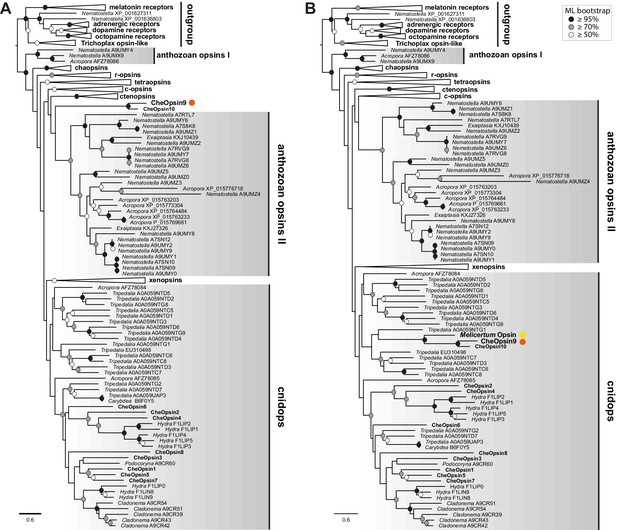
Use of the GTG+Γ amino acid substitution model on the 'source data 1’ alignment generates a Long Branch Attraction artefact.
(A) Maximum likelihood (ML) analysis of the dataset analysed in Figure 6A using the GTR+Γ model leads to Clytia Opsin9 (orange dot) and Opsin10 to cluster with the anthozoan opsin II clade (RAxML, Figure 6—source data 2). (B) Addition of a single Opsin9/10 related sequence from unpublished transcriptome data of the hydrozoan medusa Melicertum octocostatum (yellow dot) breaks the long Clytia Opsin9/10 branch and repositions them into the cnidops (RAxML, GTR+Γ, Figure 6—source data 1). ML bootstrap support values (500 replicates) are shown as coloured circles on the branch tips: black circles ≥ 95%, grey circles ≥ 70%, white circle ≥50%. Labelling as in Figure 6.

Model for Opsin9 function in Clytia oocyte maturation.
Model for Clytia oocyte maturation. At dawn, light activates Opsin9 in specialised cells of the gonad epithelium, causing secretion of MIH inside the gonad. MIH in turn acts on the oocyte surface to trigger the resumption of meiosis, followed by Germinal Vesicle breakdown (GVBD) and oocyte maturation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene (Clytia hemishaerica) | CheOpsin4 | this study | GenBank MF435920 | sequence retrieved from Clytia trancriptome data |
| gene (C. hemisphaerica) | CheOpsin7 | this study | GenBank MF435921 | sequence retrieved from Clytia trancriptome data |
| gene (C. hemisphaerica) | CheOpsin9 | this study | GenBank MF435922 | sequence retrieved from Clytia trancriptome data |
| genetic reagent (C. hemisphaerica) | Opsin9 n1_4 | this study | Mosaic F0 polyp colony | |
| antibody | anti-PRPamide | Takeda et al., 2018 | Affinity purified Rabbit polyclonal; against synthetic PRPamide | |
| antibody | anti-acetylated alpha-tubulin (rat monoclonal) | Sigma Aldrich | 6-11B-1 | (1:500) |
| antibody | anti alpha-tubulin (rat monoclonal) | Thermo Fisher Scientific | YL1/2 | (1:1000) |
| antibody | anti- ɣ-Tubulin | Sigma Aldrich | GTU-88, | (1:200) |
| antibody | Rhodamine goat anti-rabbit, Cy5 goat anti-mouse-IgG, Cy5 goat anti-rabbit- secondaries | Jackson ImmunoResearch | (1:1000) | |
| other | rhodamine phalloidin | Molecular Probes | (1:250) | |
| other | Hoechst dye 33258 | Sigma Aldrich | (1:5000) | |
| guide RNA (sgRNA) | Opsin9 n1 | this study | sequence given in Figure 3—source data 1 | |
| recombinant DNA reagent | pDR27457 (plasmid) | Addgene | 42250 | |
| Transcriptome data | Clytia hemisphaerica reference transcriptome | This study | GSE101072_Clytia_tophat_ trinityGG_combined_v2 | Combined assembled transcriptome data from multiple samples. |
| Transcriptome data | Clytia gonad tissue samples | This study | GSM2698879 GEN-BR1 GSM2698880 GEN-BR2 GSM2698881 GEC-BR1 GSM2698882 GEC-BR2 GSM2698883 GrOo-BR1 GSM2698884 GrOo-BR2 GSM2698885 FGOo-BR1 GSM2698886 FGOo-BR2 | Illlumina HiSeq 50nt reads for 8 RNA samples (four tissues, two duplicates for each tissue ) |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29555.020





