Defective STIM-mediated store operated Ca2+ entry in hepatocytes leads to metabolic dysfunction in obesity
Figures
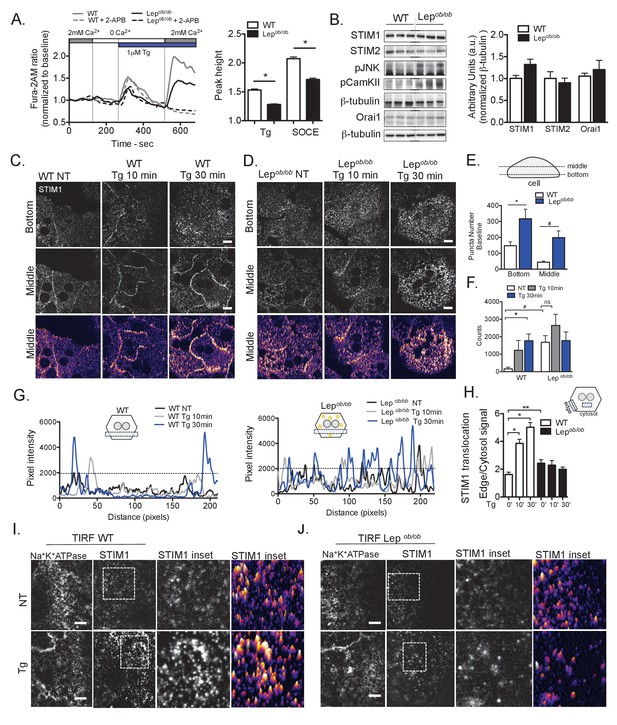
Obesity leads to decreased SOCE and impaired STIM1 translocation in primary hepatocytes.
(A) Left panel: representative Fura-2AM based cytosolic Ca2+ measurements in primary hepatocytes isolated from wild type (WT) and leptin-deficient (Lepob/ob) obese mice. ER Ca2+ stores were depleted with 1 µM thapsgargin (Tg), a SERCA inhibitor. SOCE was evaluated by substituting the extracellular media (0 mM Ca2+) with media containing 2 mM Ca2+. Dashed lines show Ca2+ measurements in the presence of 50 µM 2-Aminoethoxydiphenyl borate (2-APB). Right panel: quantification of Tg-induced Ca2+ release (reflecting ER Ca2+ content) and SOCE based on the measurements shown in (A), n = 160 WT and n = 250 Lepob/ob cells for Tg response and n = 234 WT and n = 303 Lepob/ob cells for SOCE response. Data were pooled across six independent experiments. *p<0.0001 (B) Left panel: Immunoblot analysis of protein expression levels in total liver lysate from WT and Lepob/ob mice, Right panel: quantification of the western blots n = 4, representative of 4–5 experiments. (C and D) Confocal images of immunofluorescence staining for endogenous STIM1 in primary hepatocytes from WT and Lepob/ob animals, treated with DMSO (vehicle, NT) or 1 µM Tg for 10 and 30 min. NT refers to ‘not Tg treated’ (E) Quantification of STIM1 puncta/cluster number in the bottom and middle cross-section of non-treated (DMSO) hepatocytes from WT and Lepob/ob animals.n = 5–6 fields (WT) and 4–5 fields (Lepob/ob), representative of 4 independent experiments, *p=0.02 #p=0.003 (F) Quantification of pixel intensity in a set area (125 × 125 pixels) of the middle section of the cells from WT and Lepob/ob animals, treated with 1 µM Tg or vehicle (quantification methods depicted in Figure 1—figure supplement 1 F), n = 4–9 cells, representative of 4 independent experiments, *p=0.02 #p=0.008 (G) Representative profile plots of STIM1 levels (pixel intensities) in a defined area (box) across cells treated with DMSO or 1 µM Tg for 10 or 30 min. Left: cells from WT animals, right: cells from Lepob/ob animals. (H) Quantification of STIM1 translocation by calculating the ratio between the mean STIM1 pixel intensity at a selected area of the edge of the cell relative to the same measurement performed in the cytosolic area near the edge of the cell, n = 3–4 ratios per cell, quantified in 2–8 cells for each condition *p<0.0001 **p=0.009 (I and J) Representative TIRF images of STIM1 and Na+K+-ATPase (PM marker) in cells from WT (I) and Lepob/ob mice (J) treated with 1 µM Tg or vehicle for 10 min. NT refers to “not Tg treated. For all graphs, error bars denote s.e.m. Scale: 10 µm.
-
Figure 1—source data 1
Source data for Figure 1.
- https://doi.org/10.7554/eLife.29968.004

Obesity leads to impaired STIM1 translocation in primary hepatocytes.
(A) Left panel: mRNA expression levels of indicated genes evaluated by qPCR in liver lysates from WT and Lepob/ob mice. Error bars denote s.e.m. n = 8 WT and n = 7 Lepob/ob for STIM1, n = 4 WT and n = 3 Lepob/ob for STIM2 and Orai1, *p=0.0025. Right panel: Lean and 16 weeks HFD-fed animals. n = 7 WT and Lepob/ob for STIM1, n = 3 WT and Lepob/ob for STIM2 and Orai1, *p=0.001 and #p=0.013. 18S was used as an endogenous control. (B) Left panel: Immunoblot analysis for the indicated proteins in total liver lysates from lean and 16 weeks HFD-fed animals; Right panel: protein quantifications. n = 3 Lean and n = 4 HFD. (C) Validation of STIM1 antibody for protein staining in primary hepatocytes derived from WT and STIM1- deficient animal (D) Confocal images of immunofluorescence staining for endogenous STIM1 in primary hepatocytes from WT and Lepob/ob animals, treated with DMSO (vehicle) or 1 µM Tg for 20 min. Scale: 10 um. (E) Histogram analysis of STIM1 pixel intensity measured in a defined region of the primary hepatocytes presented in Figure 1C and D, *p=0.02 (F) Schematic representation of the quantification analysis performed on confocal images shown in Figure 1C and D. For all graphs, error bars denote s.e.m.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.29968.006
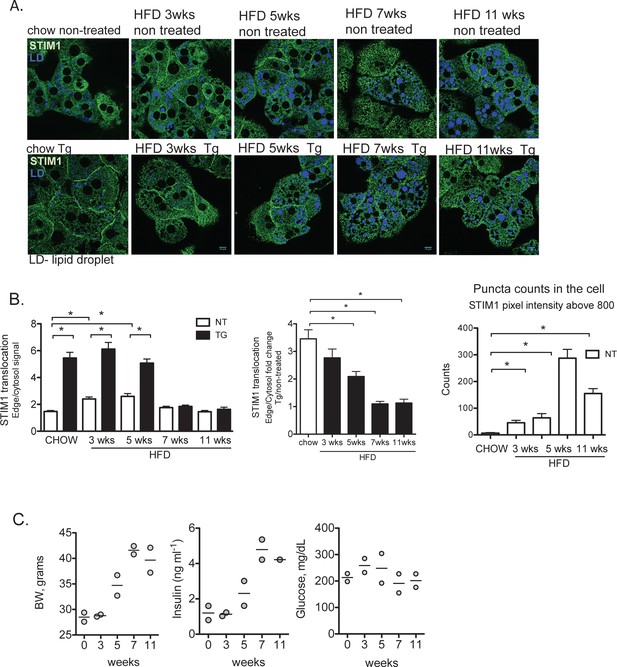
Decline of STIM1 trafficking in primary hepatocytes over the course of high fat diet (HFD)
(A) Confocal images of immunofluorescence staining for endogenous STIM1 in primary hepatocytes from lean (chow) and animals fed a HFD for 3, 5, 7 and 11 weeks following treatment with DMSO (vehicle) or 1 µM Tg for 10 min. (B) Quantification of STIM1 translocation by calculating the ratio and fold change between the mean STIM1 pixel intensity at a selected area of the edge of the cell relative to the same measurement performed in the cytosolic area near the edge. Right panel: Quantification of STIM1 puncta intensity above 800 in a cross-section of the cell. Graphs show average of 4 ratios per cell, quantified in 5 cells per field in four fields for each condition *p<0.001 (C) Body weight, fed glucose and insulin levels from animals fed chow or HFD for the indicated time. n = 2 per condition. For all graphs, error bars denote s.e.m.
-
Figure 1—figure supplement 2—source data 1
Source data for Figure 1—figure supplement 2.
- https://doi.org/10.7554/eLife.29968.008

Confocal images of immunofluorescence staining of endogenous STIM2 in primary hepatocytes from WT and Lepob/ob animals, treated with or without 1 µM Tg for 10 min.
STIM2 translocation is quantified by calculating of the edge/cytosol signal ratio. n = 10 WT NT, n = 31 WT Tg and n = 30 Lepob/ob NT and n = 43 Lepob/ob Tg, *p=0.0009, #p=0.0002.For all graphs, error bars denote s.e.m.
-
Figure 1—figure supplement 3—source data 1
Source data for Figure 1—figure supplement 3.
- https://doi.org/10.7554/eLife.29968.010
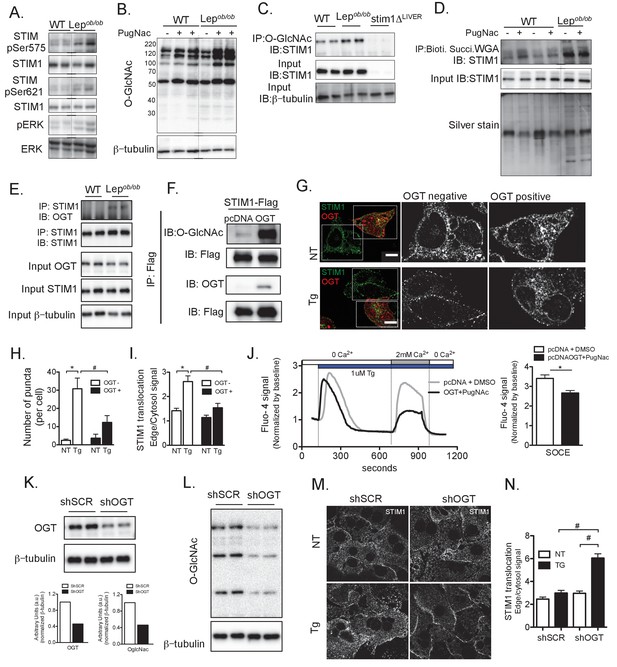
Post-translational modification of STIM1 by phosphorylation and O-GlcNAcylation in obesity.
(A) Immunoblot analysis of the phosphorylation level of STIM1 at Ser575 and Ser621 and of ERK phosphorylation in total liver lysates from WT and Lepob/ob animals (B) Immunoblot analysis of O-GlcNAcylation of total lysates extracted from primary hepatocytes derived from WT and Lepob/ob animals in the presence or absence of overnight treatment with 10 µM of PugNac, an inhibitor of O-GlcNAcase. (C) Immunoprecipitation of O-GlcNAcylated proteins derived from WT and Lepob/ob hepatocytes followed by immunoblotting for STIM1. Cells from STIM1-deficient mice (STIM1ΔLiver) were used as a negative control. (D) Total lysates from WT or Lepob/ob hepatocytes treated or not with 10 µM of PugNac overnight were incubated with biotinylated, succinylated WGA and precipitated with streptavidin-conjugated beads. The recovered proteins were used for immunoblotting for STIM1. An aliquot of each precipitate was run in a gel and silver stained. (E) Immunoprecipitation of STIM1 in total lysates from primary hepatocytes derived from WT and Lepob/ob animals followed by immunoblotting for OGT. Input for OGT and STIM1 in the total lysates prior to immunoprecipitation is shown (F) Immunoprecipitation of STIM1-Flag from total lysates stably expressing STIM-Flag in the presence or absence of OGT followed by immunoblotting for O-GlcNac and OGT. (G) Confocal images of endogenous immunofluorescent staining of STIM1 in Hepa1-6 cells transfected with OGT, treated with DMSO or 1 µM Tg stimulation. Non-transfected cells present in the same dish were used as controls for quantification. OGT expressing cells were identified by the presence of RFP expression. NT refers to ‘not Tg treated’ (H) Number of STIM1 puncta quantified per cell, n = 7–14 cells per group, representative of 3 independent experiments, *p=0.0003 #p=0.03. (I) Quantification of STIM1 translocation by calculating the ratio between STIM1 protein signal at the edge of cell relative to the cytosol close by. For OGT n = 60–64 areas and for OGT +n = 22–30 areas per group, representative of 3 independent experiments,*p<0.0001 #p=0.004 (J) Left panel: representative Fura-2AM based cytosolic Ca2+ measurements in Hepa1-6 cells overexpressing pcDNA control or OGT. Right panel: quantification of Ca2+ influx through SOCE. n = 51 (pcDNAt +DMSO) n = 61 (OGT +PugNAc) *p=0.0007. (K and L) Immunoblot analysis and densitometric quantification of OGT and OglcNAc expression in primary hepatocytes from Lepob/ob animals infected with adenovirus expressing scrambled shRNA (shSCR) and shRNA against OGT (shOGT) for 48 hr. (M) Confocal images of immunofluorescence staining for endogenous STIM1 in primary hepatocytes from Lepob/ob animals expressing scrambled shRNA (shSCR) and shRNA against OGT (shOGT) for 48 hr. NT refers to ‘not Tg treated’ (N) Quantification of STIM1 translocation by calculating the ratio between the mean STIM1 pixel intensity at a selected area of the edge of the cell relative to the same measurement performed in the cytosolic area near the edge of the cell, n = 3 ratios per cell, 5 cells per field in four fields per condition,*p=0.04 # p<0.001.For all graphs, error bars represent s.e.m. Scale: 10 µm.

STIM1 O-GlcNAcylation in primary hepatocytes and Hepa 1-6 cells.
(A) Immunoblot analysis of STIM1 from cells derived from animals on chow or HFD for 3, 5 and 11 weeks incubated with biotinylated, succinylated WGA and precipitated with streptavidin-conjugated beads. (B) Immunoblot analysis of total O-GlcNAcylation in total lysates from Hepa1-6 cells non-transfected or transfected with pcDNA empty vector or pcDNA expressing OGT (C) Immunoblot analysis of Flag and OGT in total lysates of HEK293 cells stably overexpressing STIM1-Flag in the presence or absence of overexpression of OGT.
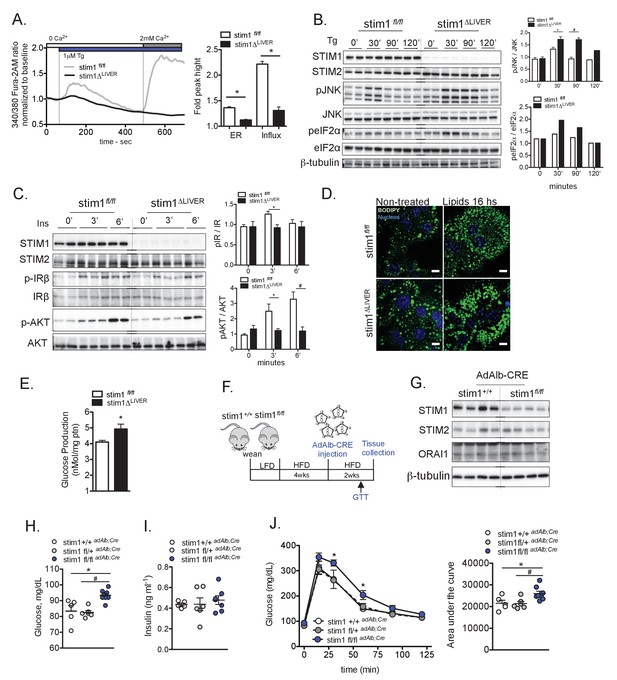
STIM1 deficiency leads to increased cellular stress, insulin resistance and lipid droplet accumulation.
(A) Left panel: representative Fura-2AM-based cytosolic Ca2+ measurements in primary hepatocytes isolated from stim1fl/fl and stim1ΔLIVER mice. Right panel: quantification of Tg-induced Ca2+ release (reflecting ER Ca2+ content) and SOCE. Error bars denote s.e.m. n = 98 WT and n = 116 stim1ΔLIVER for Tg response and n = 69 stim1fl/fl and n = 101 stim1ΔLIVER for SOCE. Cells were pooled across four independent experiments. *p<0.0001 (B) Immunoblot analysis and quantification of the expression levels of stress markers in primary hepatocytes isolated from stim1fl/fl and stim1ΔLIVER mice. n = 6 samples in each group, pooled across three independent experiments for pJNK and n = 2 for peIF2α, *p=0.0138 #p<0.0001. (C) Immunoblot analysis and quantification of insulin signaling before and after 3 nM insulin (Ins) treatment for the indicated time points in primary hepatocytes isolated from stim1fl/fl and stim1ΔLIVER mice. n = 4–5 samples in each group, pooled across two independent experiments for pIR. *p=0.011 and pAKT *p=0.03 and #p=0.0088 (D) Primary hepatocytes isolated from stim1fl/fl and stim1ΔLIVER animals treated or not with a mixture containing 1 mM Oleic Acid and 40 µM palmitic acid for 16 hr. Lipid droplets were stained in green with BODIPY and the nucleus in blue with DAPI. Scale: 10 µm. (E) Glucose output derived from primary hepatocytes isolated from stim1fl/fl and stim1ΔLIVER mice kept on a HFD, n = 4 stim1fl/fl and n = 4 stim1ΔLIVER*p=0.046 (F) Schematic representation of the protocol for adenovirus-mediated transient hepatocyte knockdown of STIM1 (G) Immunoblot analysis of the indicated proteins in total liver lysates derived from stim1+/+ n = 4 and n = 4 stim1fl/fl mice, both groups expressing liver-specific Cre recombinase (ad Alb;Cre) adenovirus. (H and I) 16 hr fasting blood glucose levels (H) and insulin (I) in animals of the indicated genotypes. *p=0.0094 stim1+/+ versus stim1fl/fl and #p<0.0001 stim1fl/+ versus stim1fl/fl (J) Glucose tolerance test (GTT) in stim1+/+, stim1fl/+ and stim1fl/fl animals expressing adenoviral Alb;Cre recombinase *p<0.04; Right panel: quantification of area under the curve from GTT. n = 5 stim1+/+, n = 6 stim1fl/+, n = 8 stim1fl/fl animals, *p=0.03 stim1+/+, versus stim1fl/fl and #p=0.01 stim1fl/+ versus stim1fl/fl, representative of 2 independent experiments. For all graphs, error bars denote s.e.m.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.29968.014

STIM1 downregulation increases cellular stress and impairs insulin signaling in Hepa 1-6 cells.
(A and B) Immunoblot analysis and quantification of expression levels of indicated proteins in Hepa 1–6 cells stably expressing shRNA against STIM1 (A) or STIM2 (B). n = 3 for SCR and STIM1 shRNA, *p=0.0005 in (A) and s.e.m. n = 3 for SCR and STIM2 shRNA, *p<0.0001 in (B). (C) Left panel: representative Fura-2AM based cytosolic Ca2+ measurements in Hepa1-6 cells stably expressing a control scrambled shRNA or shRNA against STIM1. Right panel: quantification of Tg induced Ca2+ release (reflecting ER Ca2+ content) and SOCE. n = 79–92 cells in each group, pooled across five independent experiments. *p<0.0001 (D) Left panel: Immunoblot analysis of indicated proteins in total lysates of Hepa 1–6 cells stably expressing scrambled shRNA or shRNA against STIM1 treated with DMSO or 100 nM thapsgargin (Tg) for the indicated time points. Right panel: quantification of Immunoblots. n = 4 shRNA SCR and shRNA STIM1, *p=0.04 #p=0.02 for pJNK and *p=0.03 for peIF2. (E) Immunoblot analysis of indicated proteins in total lysates Hepa 1–6 cells stably expressing scrambled shRNA or shRNA against STIM2 treated with DMSO or 100 nM thapsgargin (F) Immunoblot analysis of expression levels of markers of insulin signaling in total lysates of Hepa 1–6 cells expressing shScramble and shSTIM1 treated with 3 nM insulin (Ins) treatment for the indicated time points. (G) Immunoblot analysis of expression levels of markers of insulin signaling in total lysates of Hepa 1–6 cells expressing shScramble and shSTIM1 treated with 3 nM insulin (Ins) treatment for the indicated time points. For al graphs, error bars denote s.e.m.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.29968.016

Effect of liver-specific deletion of STIM1 on metabolic parameters in vivo.
(A) Right panel: Immunoblot analysis of expression levels of indicated proteins in liver lysates from of stim1fl/fl and STIM1 hepatocyte-specific knockout stim1ΔLIVER. Left panel: Quantification of immunoblots. n = 5 stim1fl/fl, n = 3 stim1fl/+, n = 5 stim1ΔLIVER, *p<0.001 #p<0.0057. (B) Weight gain curves of stim1fl/fl and stim1ΔLIVER on chow (n = 5 stim1fl/fl, n = 8 stim1ΔLIVER) or HFD (n = 21 stim1fl/fl, n = 6 stim1ΔLIVER). Representative of 2 independent experimental cohorts. *p=0.02. (C) Glucose tolerance test at 6 weeks on HFD, n = 10 stim1fl/fl, and n = 7 stim1ΔLIVER (D) Markers of in vivo insulin signaling evaluated by immunoblot analysis of total liver lysates from animals injected with insulin (0.75 U/kg) through the portal vein. Tissues were collected 3 min after injection. Right panel: phospho-protein level quantification normalized to total protein levels. n = 3 for stim1fl/fl and n = 6 for stim1ΔLIVER, *p=0.02. (E) Liver triglyceride content in stim1fl/fl n = 11 and n = 7 stim1ΔLIVERn = 10 on 6 weeks HFD. (F) Glucose tolerance test at 20 weeks on HFD, n = 11 stim1fl/fl, and n = 16 stim1ΔLIVER representative of 2 independent cohorts. (G) Markers of in vivo insulin signaling performed in similar conditions as in D. (H) Liver triglyceride content in stim1fl/fl n = 6 and stim1ΔLIVERn = 6 on 20 weeks HFD. (I) Quantification of micro and macrovesicular steatosis in liver sections stained with Hematoxylin and Eosin. stim1fl/fl n = 6 stim1ΔLIVERn = 4, p=0.01. For all graphs, error bars denote s.e.m.
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2.
- https://doi.org/10.7554/eLife.29968.018
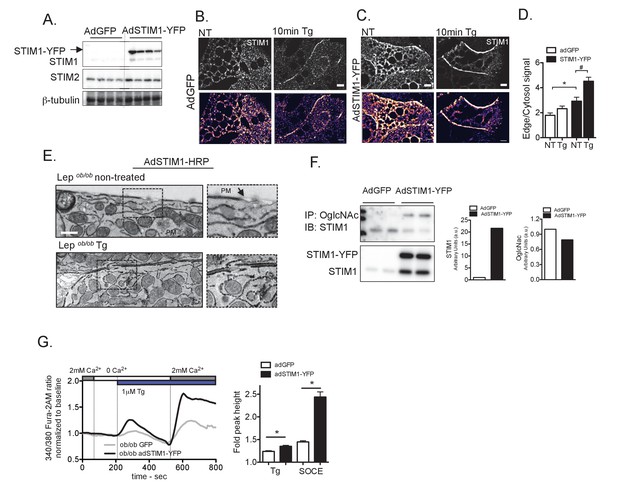
Overexpression of STIM1 promotes STIM1 translocation to ER/PM junctions and improves SOCE in primary hepatocytes from obese animals.
(A) Immunoblot analysis of STIM1-YFP expression levels in total lysates from livers isolated from Lepob/ob mice transduced with adenovirus (Ad) expressing GFP or STIM1-YFP. (B–C) Confocal images of immunofluorescence staining for endogenous STIM1 in primary hepatocytes from Lepob/ob mice expressing adGFP (B) or adSTIM1-YFP (C) treated with DMSO (vehicle) and 1 µM Tg. NT refers to ‘not Tg treated’. (D) Quantification of STIM1 translocation by calculating the ratio between the mean STIM1 pixel intensity at a selected area of the edge of the cell relative to the same measurement performed in the cytosol. n = 26 adGFP NT and Tg treated and n = 16 adSTIM1-YFP NT and n = 19 Tg treated *p=0.0023, #p=0.0015. Scale: 10 µm. (E) Transmission Electron Micrographs of primary hepatocytes isolated from Lepob/ob animals expressing STIM1-HRP, treated with DMSO or treated 1 µM Tg for 10 min. Scale: 500 nm. (F) Immunoblot analysis and densitometric quantification of OglcNac and STIM1 expression in primary hepatocytes from Lepob/ob animals infected with AdGFP and AdSTIM1-YFP adenovirus for 24 hr. Quantification reflects the sum of the endogenous and exogenous STIM1 (G) Left: Representative Fura-2AM-based cytosolic Ca2+ measurements in primary hepatocytes isolated from Lepob/ob animals expressing adGFP or adSTIM1-YFP. Right: Quantification of Tg-induced Ca2+ release (reflecting ER Ca2+ content) and SOCE measurements . n = 126 adGFP and n = 83 adSTIM1-YFP cells for Tg response and n = 127 adGFP and n = 62 adSTIM1-YFP cells for SOCE, representative of 5 independent experiments. *p<0.0001. For all graphs. error bars denote s.e.m.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.29968.020

Effect of STIM1-YFP overexpression in ER morphology and apposition to the plasma membrane.
(A) Transmission Electron Micrographs of primary hepatocytes isolated from Lepob/ob animals, expressing adGFP or adSTIM1-YFP.
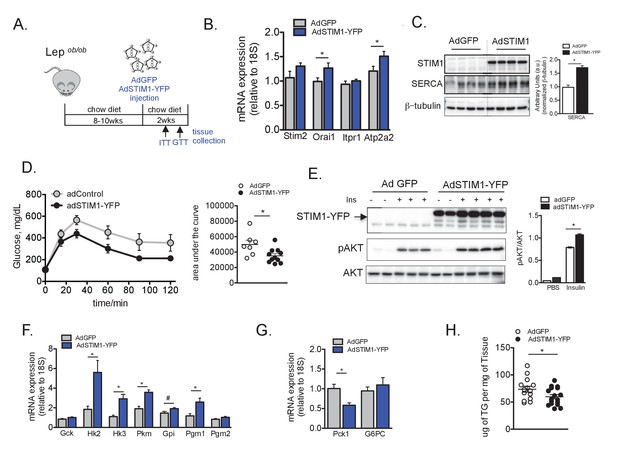
Overexpression of STIM1 leads to improved insulin signaling, glucose tolerance and lipid accumulation in obese animals.
(A) Schematic representation of the protocol for adenovirus mediated STIM1- YFP expression in Lepob/ob mice (B) mRNA expression levels of indicated genes evaluated by qPCR, normalized to 18S, n = 9 for WT and n = 10 for STIM1-YFP pooled from two independent experiments, *p=0.04 (C) Left panel: Immunoblot analysis of indicated proteins in liver total lysates from Lepob/ob mice expressing GFP or STIM1-YFP. Right panel: quantifications of the blots. n = 3 adGFP and n = 4 adSTIM1-YFP, *p=0.001 (D) Glucose tolerance tests in Lepob/ob mice after adGFP (control) and adSTIM1-YFP expression with quantifications of area under the curve. n = 7 adGFP and n = 11 adSTIM1-YFP animals, representative of 3 independent experiments.*p=0.022 (E) Markers of in vivo insulin signaling evaluated by immunoblot analysis of total liver lysates from animals injected with insulin (0.75 U/kg) through the portal vein. Tissues were collected 3 min after injection. Right panel: phospho-protein level quantification normalized to total protein levels. n = 3 for adGFP and n = 4 for adSTIM1-YFP, *p=0.0003. (F and G) mRNA expression levels of indicated genes evaluated by qPCR, normalized to 18S, n = 9 for both WT and STIM1-YFP pooled from two independent experiments. *p<0.01 # p<0.04. (H) Liver triglyceride content in Lepob/ob animals expressing adGFP (control) and adSTIM1-YFP pooled from three independent cohorts. n = 14 adGFP and 17 adSTIM1-YFP, *p=0.042. For all graphs, error bars denote s.e.m.
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.29968.024
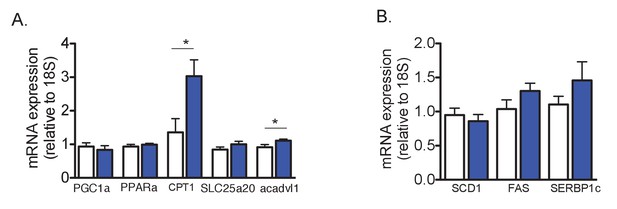
Effect of STIM1-YFP overexpression in mRNA expression of genes involved in lipolysis and lipogenesis.
(A and B) mRNA expression levels of indicated genes evaluated by qPCR, normalized to 18S, n = 8 for WT and n = 9 for STIM1-YFP pooled from two independent experiments, *p<0.02.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://doi.org/10.7554/eLife.29968.026
Videos
STIM1-YFP translocation upon thapsigargin (Tg) treatment.
Primary Hepatocytes were infected with STIM1-YFP, treated with 1 uM Tg and YFP fluoresce was recorded over the period of time indicated in the movie.
Tables
List of primers used for qPCR measurements and sequences of the shRNA used in this study.
https://doi.org/10.7554/eLife.29968.027| List of SYBR green primers for real-time PCR | ||
|---|---|---|
| Gene name | Forward primer | Reverse primer |
| stim1 | TGAAGAGTCTACCGAAGCAGA | AGGTGCTATGTTTCACTGTTGG |
| stim1 | ACAGTGAAACATAGCACCTTCC | TCAGTACAGTCCCTGTCATGG |
| stim2 | CGAAGTGGACGAGAGTGATGA | GGAGTGTTGTTCCCTTCACATT |
| orai1 | GATCGGCCAGAGTTACTCCG | TGGGTAGTCATGGTCTGTGTC |
| Itpr1 | GGGTCCTGCTCCACTTGAC | CCACATCTTGGCTAGTAACCAG |
| Atp2a2 | CTGTGGAGACCCTTGGTTGT | CAGAGCACAGATGGTGGCTA |
| Gck | ACCAAGCGGTATCAGCATGTG | TGGACTTCTCTGTGATTGGCA |
| Hk2 | ATGATCGCCTGCTTATTCACG | CGCCTAGAAATCTCCAGAAGGG |
| Hk3 | TGCTGCCCACATACGTGAG | GCCTGTCAGTGTTACCCACAA |
| Pkm | GGTGGCTCTGGATACAAAGGG | CACACTTCTCCATGTAAGCGT |
| Gpi | CTCAAGCTGCGCGAACTTTTT | GGTTCTTGGAGTAGTCCACCAG |
| Pgm1 | CAGAACCCTTTAACCTCTGAGTC | TCATTCATTCGAGAAATCCCTGC |
| Pgm2 | GCGGAATGGGATGAACAAGGA | GGTCATTGATGTAGCAAAACCCT |
| Pck1 | CTGCATAACGGTCTGGACTTC | GCCTTCCACGAACTTCCTCAC |
| G6pc | CTGAGCGCGGGCATCATAAT | GATTCTTAGGATCGCCCAGAAAG |
| Pgc1a | CCC TGC CAT TGT TAA GAC C | TGC TGC TGT TCC TGT TTT C |
| Ppara | TATTCGGCTGAAGCTGGTGTA | CTGGCATTTGTTCCGGTTCT |
| Cpt1 | GCTGGAGGTGGCTTTGGT | GCTTGGCGGATGTGGTTC |
| Slc25a20 | AGTCGGACCTTGACCGTGT | GACGAGCCGAAACCCATCAG |
| Scd1 | TTC TTG CGA TAC ACT CTG GTG C | CGG GAT TGA ATG TTC TTG TCG T |
| Fas | GGA GGT GGT GATA GCC GG TAT | TGG GTA ATC CATA GAG CCC AG |
| Serbpc1 | GGAGCCATGGATTGCACATT | GGCCCGGGAAGTCACTGT |
| 18S | AGTCCCTGCCCTTTGTACACA | CGATCCGAGGGCCTCACTA |
| Oligonucleotides for shRNA | ||
| stim1 | CCGGCCGAAACATCCATAAGCTGATCTCGA GATCAGCTTATGGATGTTTCGGTTTTTTG | TRCN0000193877 |
| stim2 | CCGGGACGAAGTAGACCACAAGATTCTCG AGAATCTTGTGGTCTACTTCGTCTTTTTTG | TRCN0000187841 |
| ogt | CCGGCCCATTTCTTTCAGCAGAAATCTCGA GATTTCTGCTGAAAGAAATGGGTTTTTG | TRCN0000110395 |
List of antibodies used in this study.
https://doi.org/10.7554/eLife.29968.028| Antibodies used in western blots | ||
|---|---|---|
| Antibody | Vendor | Catalog number |
| STIM1 | Cell signaling | #4916 |
| STIM2 | Cell signaling | #4917 |
| ORAI1 | Sigma | SAB3500126 |
| SERCA2b | Cell Signaling | #4388 |
| pJNK | Cell signaling | #81E11 |
| peIF2alpha | Cell signaling | #3597 |
| CHOP | Cell Signaling | mab# 2895 |
| pAKT | Santa Cruz | 7985R |
| AKT | Santa Cruz | 8312 |
| IR | Santa Cruz | 711 |
| pIR | Calbiochem | 407707 |
| OGT | Cell Signaling | #5368 |
| O-GlcNAc (CTD110.6) | Cell Signaling | #12938 |
| Flag M2 | Sigma | SLBD9930 |
| pCamKII | Cell Signaling | #12716 |
| β-tubulin | Abcam | ab 21058 |
| pSTIM1 | provided by Dr. Martin-Romero, University of Extremadura, Spain | |
Additional files
-
Supplementary file 1
Source uncropped western blots: this file contains all the un-cropped western blot images presented in this manuscript.
- https://doi.org/10.7554/eLife.29968.029
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29968.030





