Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae
Figures
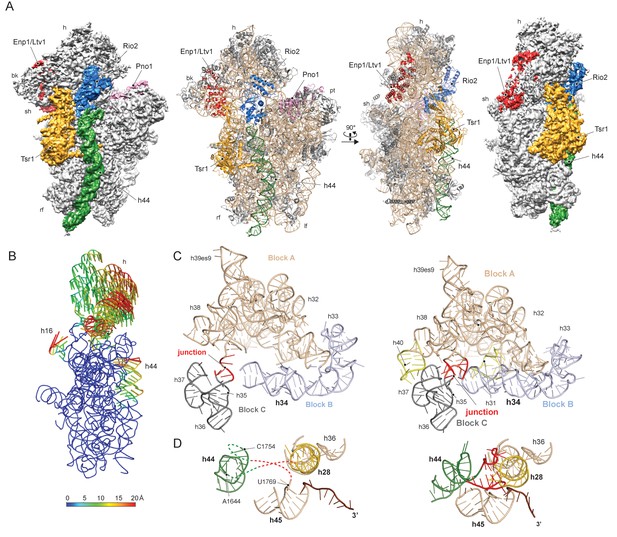
Structure of the pre-40S particle and conformation of the 18S rRNA..
(A) 3.6 Å cryo-EM reconstruction and molecular model of the pre-40S complex containing Enp1/Ltv1, Tsr1, Rio2 and Pno1 (B) Conformational transition of the 18S rRNA from the pre-mature to the mature state represented by vectors (superimposed on the 40S body). (C) Condensation of the head-forming rRNA on the h34/35/38 three-way junction from pre- (left) to mature state (right). Block A (h29–h32, h38–h42) mostly resembles the mature state and served as the moiety for superposition. Relative to block A, blocks B (h34 and 33) and C (h35–h37) are still shifted, since the three-way junction linking the blocks is not yet established. The connection of h40 to h37 and the loop of h31 are not established in the pre-state. (D) View focusing on the the linker between h44 with h28 and h45 from the central region of 18S rRNA. In the pre-state (left).he linker region as well as parts of h44 are unfolded and h28 (yellow) is tilted compared to the mature state (right).
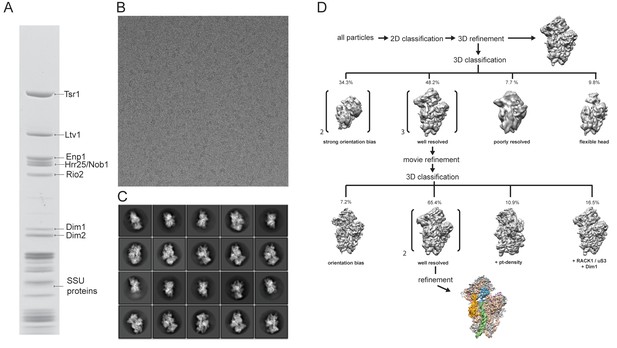
Purification and cryo-EM of the pre-40S complex.
(A) Tandem affinity purification of pre-40S from yeast using Ltv1-FLAG-Tev-ProtA as bait. Final eluates were analysed by SDS-PAGE and Coomassie staining. (B) Representative micrograph showing pre-40S·particles. (C) Representative 2D classes showing various projections of pre-40S subunits. (D) Classification scheme. 2D classification was used to remove non-ribosomal particles. After refinement and 3D classification in RELION-2 (see Materials and methods) three major classes were obtained. The main class showed density for the RBFs Tsr1, Enp1/Ltv1, Rio2 and Pno1 and was refined to 3.6 Å. Two other classes showed extra densities not seen in the main class. One class displayed distorted density at the platform in a position previously attributed to Nob1 (Strunk et al., 2011). The other class showed density for RACK1 and parts of uS3 in addition to density at a position previously attributed to Dim1 (Johnson et al., 2017). However, due to the small number of particles and orientation bias, these classes were not further refined.
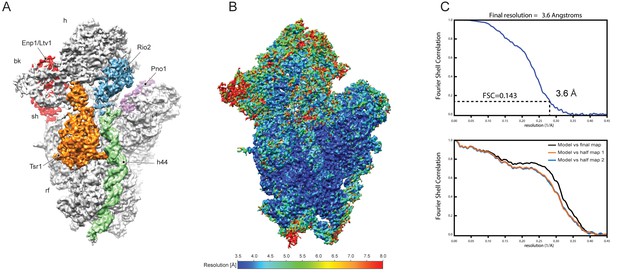
Assessment of resolution and model quality of the cryo-EM structure.
(A) Cryo-EM density of the pre-40S complex low-pass filtered at 3.6 Å showing the 40S subunit (grey), Tsr1 (orange), Enp1 (red), Rio2 (blue) and Pno1 (pink). The rRNA helix h44 is highlighted in green. (B) Local resolution of the pre-40S complex as calculated by ResMap (Kucukelbir et al., 2014). The ResMap plots show a range from a maximum of 3.5 Å to 8.0 Å in the periphery. (C) Fourier Shell Correlation (FSC) plot showing the 3.6 Å average resolution of the map according to the ‘gold standard’ criterion (FSC = 0.143; top) and FSC curves calculated between the cryo-EM map and the final model (bottom) as calculated by REFMAC. Values are plotted for the model versus the final map (FSC average, black), for the model that was refined into the first half-map and FSC calculated either for the same map (model vs first half-map, orange) or for the second half-map (model vs second half map, blue).
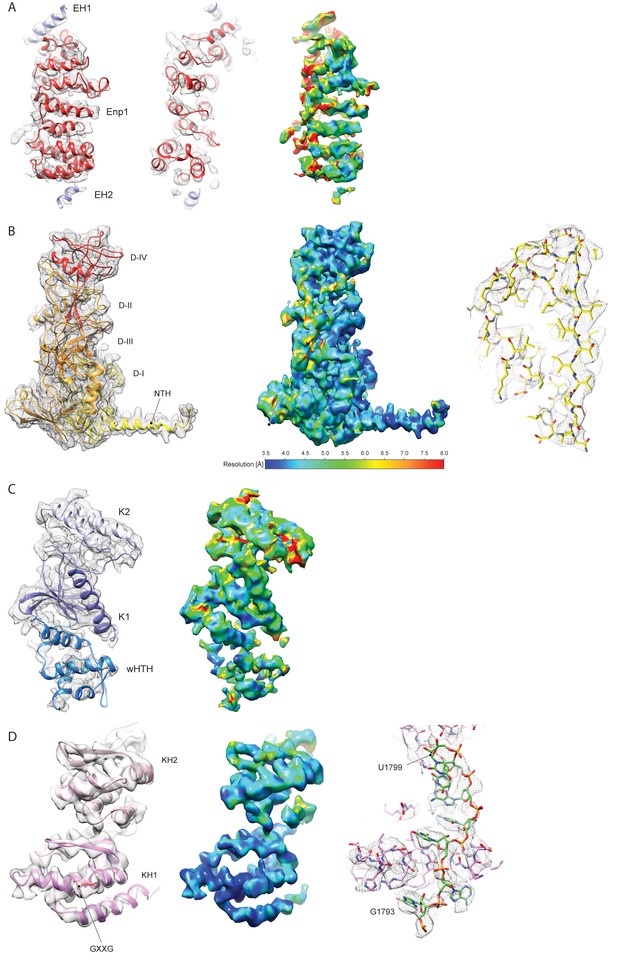
Fitting of the RBFs.
(A) Model for Enp1 fitted into isolated density low-pass filtered at approximately 5 Å. The model for Enp1 is shown in red, α-helical extra density was observed at the N- and C-terminal regions of Enp1 and the model built as poly-Ala (EH1 and EH2; blue). These extra densities could be attributed to of Enp1 or to the bait protein Ltv1. At the given local resolution (between 4.5 and 8.5 Å) no molecular details are present to allow unambiguous assignment of the helices to either factor. (B) Model for Tsr1 fit into the isolated density low-pass filtered at 3.6 Å. A high local resolution allowed unambiguous identification of most sidechains as exemplified on the right (zoom into domain I) and de novo determination of the α-helical structure in the Tsr1 N-terminus (see Figure 3a). (C) Model for Rio2 fit into isolated density low-pass filtered at 5 Å. At a local resolution ranging mostly from 4 to 7.5 Å, all domains could be unambiguously docked. However, most of the loop regions displayed flexibility and could not be modelled at a molecular level. (D) Model for Pno1 fit into isolated density low-pass filtered at 3.6 Å. High local resolution below 3.5 Å, especially for KH2, allowed the modelling of most side chains and contacts to the 3’ end of 18S rRNA as depicted on the right.
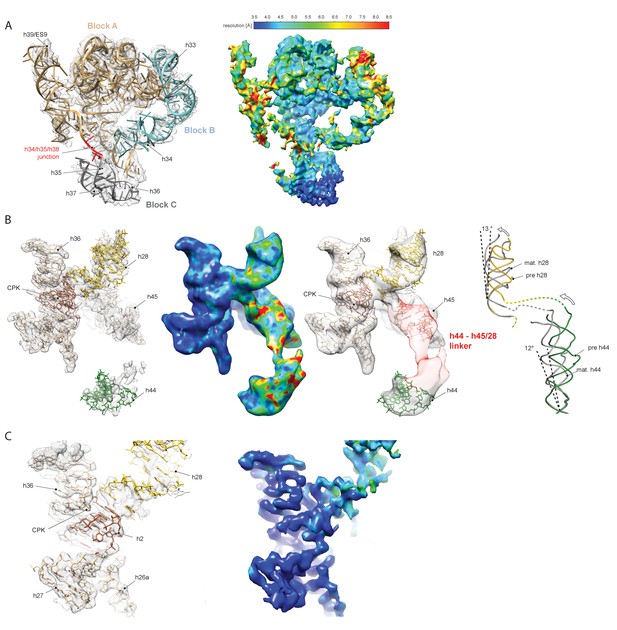
Conformation and flexibility of the pre-18S rRNA
(A) Model for the head-forming 3’ major domain (3’M; h28–h43) fitted into the cryo-EM density (left). The 3’M is divided into three blocks (block A, h29-h32; h38-h44, wheat; block B, h33-h34, light blue; block C, h35-h37, grey). The local resolution for block C and the major parts of the block A (h29-h32/h41-h43) is between 3.5 and 5.5 Å consistent with a stable and mature fold of these domains, whereas h39 and h40 and block B (h33–h34) are rather flexible, as indicated by a lower local resolution (right). Maps are low-pass filtered at 3.6 Å. (B) Model, density and local resolution of the central region of 18S rRNA connecting the 5’- (h1 and h2), central (h2 and h27), 3’M (h36) and the 3’m (h44) domains (left). h1 and h2 form the central pseudoknot (CPK; brown). While the 5’- and central domains as well as h36 form a rigid structure as indicated by high local resolution (middle left), h28 and the 3’m are more flexible and the entire linker region connecting h44 with h28 and h45 is not visible when low-pass filtered at 3.6 Å (left). Density for these flexible nucleotides (1633–1644 and 1754–1769; red) is only visible when low-pass filtering the map to below 15 Å (middle right). When compared to the mature state, h28 and h44 need to undergo a conformational transition. H28 is kinked by 13 degrees relative to its mature position and h44 is rotated outward towards the solvent side by 12 degrees (right). (C) view focusing on the rigid region around the CPK fitted into electron density (left). The connection between the 5’ and central domains to h36 of 3’M is already stably formed in the pre-40S particle as indicated by high local resolution below 3.5 Å, whereas h28 is more flexible (right).
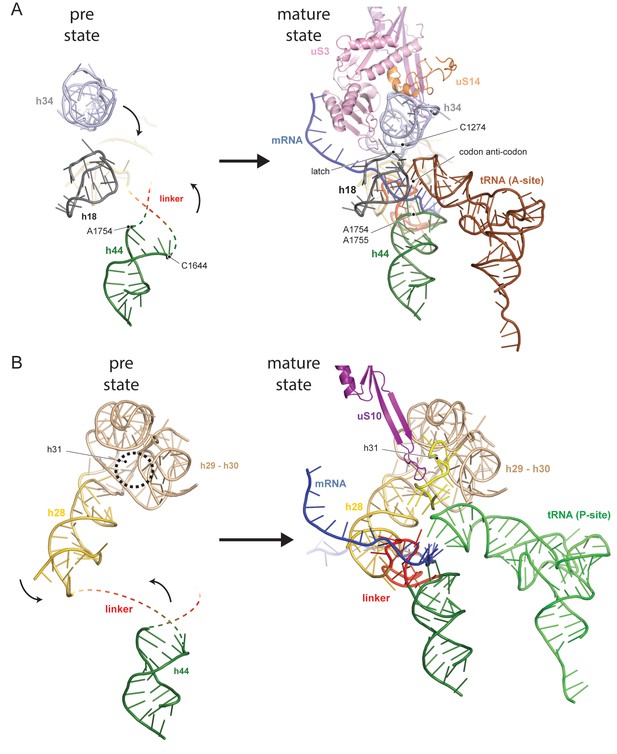
Comparison between pre- and native 40S focusing on the active sites.
(A) View focusing on the mRNA entry and A-site. The A-site is composed of h18, h34 and h44, where in a translating ribosome the anticodon-loop of a A-tRNA is bound and the mRNA enters the 40S via the latch structure formed between h18, h34 and uS3. In the pre-40S, uS3 is absent and h34 is displaced. Moreover, h44 is shifted and its tip including the decoding adenines A1754 and A1755 is unfolded. (B) View focusing on the mRNA exit and the P-site. The P-site is composed of h24 (not shown), h28, h29 and h31 as well as the linker between h44 and h45. In the pre-40S this linker is delocalized and h28 is shifted. Moreover, the tip of h31 which is stabilized by uS10 in the mature state and binds the P-site tRNA in a translating ribosome is not folded in the pre-state.
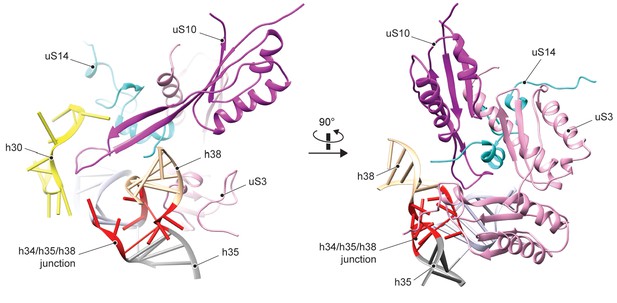
Structure and environment of the native h34-h35-h38 three-way junction.
In the mature 40S subunit, h34-h35-h38 forms a family-A three-way junction that is stabilized by r-proteins uS3, uS10 and uS14, where uS10 also stabilizes the tip of h30 (nucleotides 1189–1200). All three proteins are absent in the pre-40S particle and neither the three-way junction nor the h30 tip is formed.
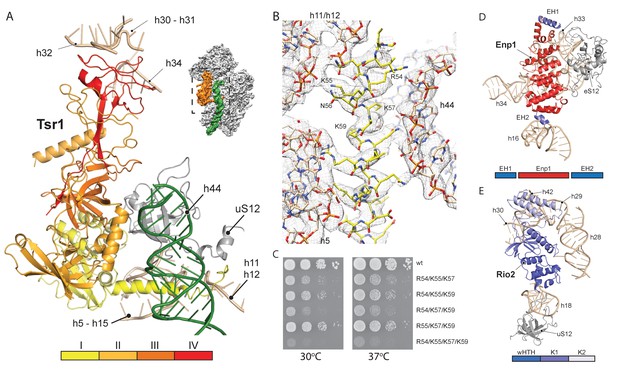
Interactions of Tsr1, Enp1/Ltv1 and Rio2 with the pre-40S.
(A) Tsr1 binds the 40S body (h5–h15, h17 and uS12) via domains I and III, the 40S head (h30–h31, h32 and h34) via domain IV. The N-terminal α-helix of Tsr1 intercalates between h44, h5 and h11-h12. (B) Model of the Tsr1 N-terminal α-helixfitted into density low-pass filtered at 3.6 Å. (C) Growth analysis of wt Tsr1 and reverse-charge point mutations in residues interacting with h44 (R54D, K55D, K57D and K59D). Constructs were transformed into a Tsr1 shuffle strain and selected on SDC + FOA plates. Strains were spotted in 10-fold serial dilution on YPD plates and incubated for 2 days at the indicated temperatures. Different temperatures were used to assess if the growth defect observed at 30 degrees was enhanced at higher temperatures. (D) Enp1 binds to h33, h34 and eS12 and to the kinked h16; EH = extra helix for Enp1 or Ltv1. (E) Rio2 binds the 40S body via its N-terminal winged-helix-turn-helix-domain (wHTH) (h18, uS12), and the 40S head via the two-lobed kinase domain (K1/K2) (h28,h30). Moreover, K2 reaches into the P-site contacting h29, h42 and the region in between.
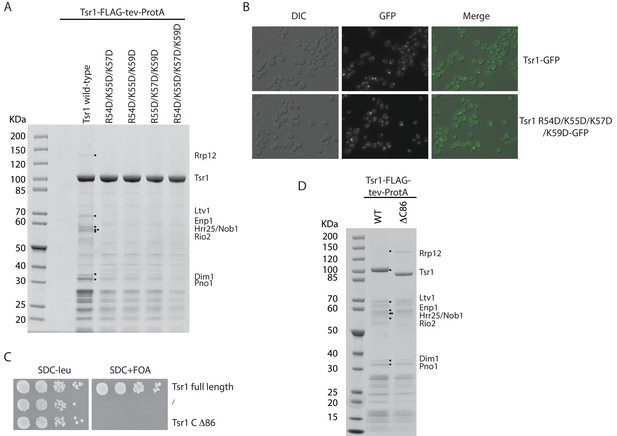
Functional analyses of mutant forms of Tsr1.
(A) Tandem affinity purification of WT Tsr1 (Tsr1-FTpA) or the indicated mutants harbouring reverse-charge point mutations in residues interacting with h44 (R54D, K55D, K57D and K59D). Final eluates were analysed by SDS-PAGE and Coomassie staining. (B) Localization of wt Tsr1 (Tsr1-GFP) and Tsr1 harbouring reverse-charge point mutations in residues interacting with h44 (R54D, K55D, K57D and K59D). (C) Growth analysis of wt and truncated Tsr1, lacking domain IV (C∆86). Constructs were transformed into a Tsr1 shuffle strain, spotted in 10-fold serial dilution and grown on SDC-leu (2 days) and SDC + FOA (4 days) plates at 30°C. (D) Tandem affinity purification of wt Tsr1 (Tsr1-FTpA) and Tsr1 lacking domain IV (C∆86) as bait. Purifications were performed using FTpA constructs expressed from a plasmid, in a WT background. Final eluates were analysed by SDS-PAGE and Coomassie staining.
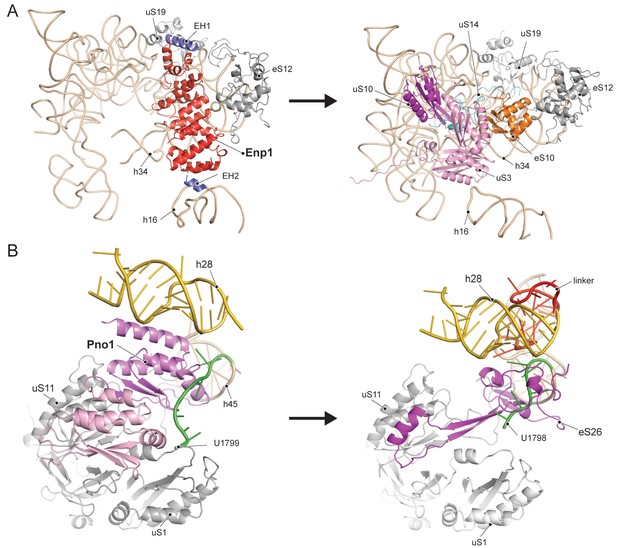
Comparison of RBF binding sites in the premature and the mature 40S.
(A) Enp1/Ltv1 binds near the mRNA entry channel and connects the shoulder (h16) with the beak (h33, h34 and eS12). It occupies the position of the eS10. Moreover, uS3, uS10 and uS14 are not incorporated into the pre-40S particle. Note that h16 is in a bent conformation compared to the mature state. (B) Pno1 binds at the platform of the pre-40S contacting uS1, uS11, the kinked h28 as well as h45 and the 3’-end of 18S rRNA. It thereby occupies the position of eS26, which binds the rearranged 3’ end in the mature state.
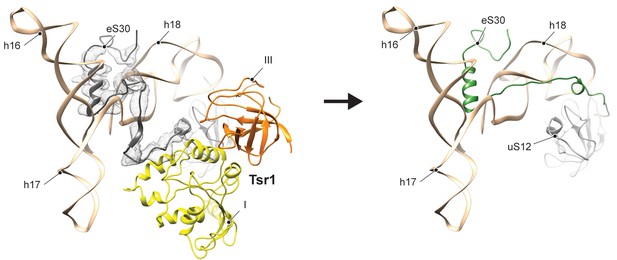
Binding of Tsr1 to h17 and eS30.
Distinct contacts are formed between domain I of Tsr1 and h17 as well as eS30 (left; model shown in density low-pass filtered according to local resolution using RELION-2). Notably the eS30 N-terminus is kept in an immature position by Tsr1. In the mature 40S (right), the eS30 N-terminus flips towards uS12, but in the pre-state this contact is hampered by Tsr1 domain III.
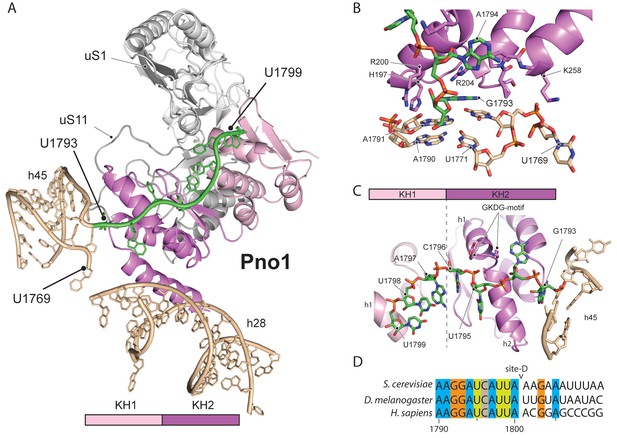
Molecular interactions of Pno1.
(A) Pno1 binds uS11/uS1, h45, the tilted h28 and the 3’ end (green) at the platform. (B) View focusing on the molecular interactions of KH2 with h45. Three C-terminal α-helices recognize the stem of h45 (A1791:U1770 and A1790:U1769), upon which G1793 is stacked. Specifically, Arg204 contacts G1793 and Arg200 together with His197 the backbone of the 3’ strand (A1791 and A1790). The loop between two of the C-terminal helices of Pno1 (Gly253-Lys258) contacts U1770 and U1769 of the 5’-strand of h45. This loop also participates in binding of the first single-stranded base A1794 which is sandwiched between Pro256 and the GKDG-loop of KH2 (C) View focusing on the molecular interactions between Pno1 and the 3’ rRNA end. Both KH domains interact with the 3’-end rRNA residues (G1793-U1799) which lead up to the D-cleavage site. The GKDG-loop of KH2 positions four nucleobases (U1795-U1798) close to its hydrophobic pocket and KH1 contacts the terminal bases (A1797-U1799) via h1 (D) Sequence alignment for eukaryotic 3’ rRNA ends showing conservation up to the D-cleavage site.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (Saccharomyces cerevisiae) | Ltv1-FTpA;Tsr1shuffle | This paper | NA | Ltv1-FTpA- Genomic copy of Ltv1 tagged at the c-terminus with FLAG-TEV-proteinA tag; Tsr1 shuffle- genomic copy of Tsr1 deleted and rescued with a plasmid with a wild-type copy |
| genetic reagent (plasmids used for expression in Saccharomyces cerevisiae) | YCplac111-Tsr1-FTpA; YCplac111-Tsr1 R54D, K55D,K57D-FTpA; YCplac111-Tsr1 R54D, K55D,K59D-FTpA; YCplac111-Tsr1 R54D, K57D,K59D-FTpA; YCplac111-Tsr1 K55D, K57D,K59D-FTpA; YCplac111-Tsr1 R54D, K55D,K57D,K59D-FTpA; YCplac111-Tsr1-GFP; YCplac111-Tsr1 R54D, K55D,K57D,K59D-GFP; YCplac111- Tsr1DC86-FTpA | This paper | NA | Plasmids containing wild-type Tsr1 or the described point mutants. Expression is under the control of the Tsr1 promoter, and are tagged at the c-terminus with either FLAG-TEV-proteinA or GFP. |
| software, algorithm | EM-TOOLS | TVIPS GmbH | NA | http://www.tvips.com/imagingsoftware/em-tools/ |
| software, algorithm | MotionCorr2.1 | https://doi.org/10.1038/nmeth.4193 | NA | http://cryoem.ucsf.edu/software/driftcorr.html |
| software, algorithm | GCTF | https://doi.org/10.1016/j.jsb.2015.11.003 | NA | http://www.mrclmb.cam.ac.uk/kzhang |
| software, algorithm | Gautomatch | public | NA | http://www.mrclmb.cam.ac.uk/kzhang |
| software, algorithm | Relion-2 | https://doi.org/10.7554/eLife.18722 | NA | http://www2.mrclmb.cam.ac.uk/relion/index.php |
| software, algorithm | Phenix suite (phenix.real_space_ refine, molprobity) | Python-based Hierarchical ENvironment for Integrated Xtallography | RRID:SCR_014224 | https://www.phenix-online.org/ |
| software, algorithm | CCP4 (LIBG, ProSMART, Refmac5, COOT) | Collaborative Computational Project No. 4 Software for Macromolecular X-Ray Crystallography | RRID:SCR_007255 | http://www.ccp4.ac.uk/ |
| software, algorithm | UCSF Chimera | UCSF Resource for Biocomputing, Visualization, and Bioinformatics | RRID:SCR_004097 | http://www.cgl.ucsf.edu/chimera/ |
| software, algorithm | Pymol | PyMOL Molecular Graphics System, Schrödinger, LLC | RRID:SCR_000305 | https://pymol.org/ |
EM data collection and refinement statistics.
https://doi.org/10.7554/eLife.30189.014| Data collection | |
|---|---|
| Particles | 84100 |
| Pixel size (Å) | 1.084 |
| Defocus range (µm) | 0.8-2.4 |
| Voltage (kV) | 300 |
| Electron dose (e- Å−2) | 28 |
| MODEL REFINEMENT | Pre40S particle |
| Model composition | |
| Non-hydrogen atoms | 71923 |
| Protein residues | 4718 |
| RNA bases | 1635 |
| Refinement | |
| Resolution for refinement (Å) | 3.7 |
| Map sharpening B-factor (Å2) | −92.8 |
| Average B-factor (Å2) | 164.6 |
| FSCaverage | 0.85 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0177 |
| Bond angles (°) | 1.61 |
| VALIDATION and STATISTICS | Pre40S particle |
| Validation | |
| Molprobity score | 2.20 |
| Clashscore, all atoms | 9.60 |
| Good rotamers (%) | 94.04 |
| Ramachandran Plot | |
| Favored (%) | 85.79 |
| Outliers (%) | 1.21 |
| Validation (RNA) | |
| Correct sugar puckers (%) | 97.8 |
| Good backbone conformations (%) | 65.6 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30189.015





