Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123
Figures
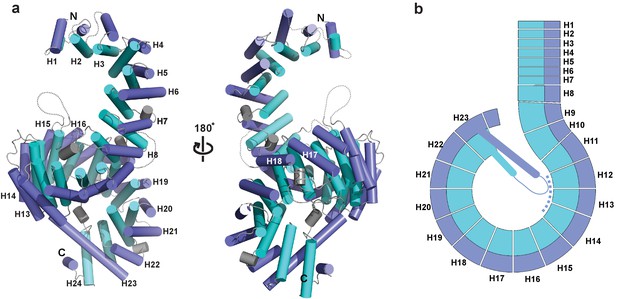
Crystal structure of full-length Kluyveromyces lactis (Kl) Kap123.
(a) The apo structure of full-length Kl Kap123 with two lateral views (180° rotation). The inner (cyan) and outer (blue) helices make up the 24 Kap123 HEAT repeats (H1–H24). The HEAT repeats form a right-handed superhelical solenoid structure with several linker regions (gray). (b) Schematic view of full-length Kl Kap123. The extra-long helix of repeat 23 forms an intramolecular interaction with repeats 12–14. The illustration incorporated in all figures was generated using PYMOL (Delano Scientific, LLC).
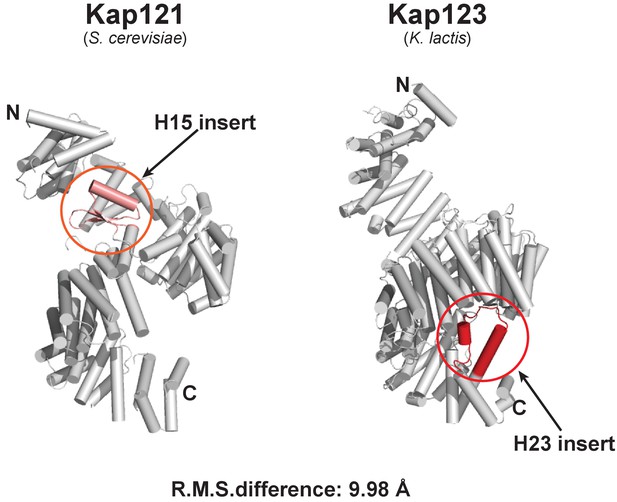
Structural comparison of full-length S. cerevisiae Kap121 and full-length K. lactis Kap123.
Cartoon side-views of S. cerevisiae Kap121 (PDB ID: 3W3T) (Kobayashi and Matsuura, 2013) and K. lactis Kap123. Although both structures display a similar architecture with the right-handed superhelical solenoid structure, each structure also possesses its own unique features. The Kap 121 H15 insert (pink) and Kap 123 H23 (red) may play important roles in substrate recognition. The root-mean-square (R.M.S.) difference of two structures is 9.98 Å.
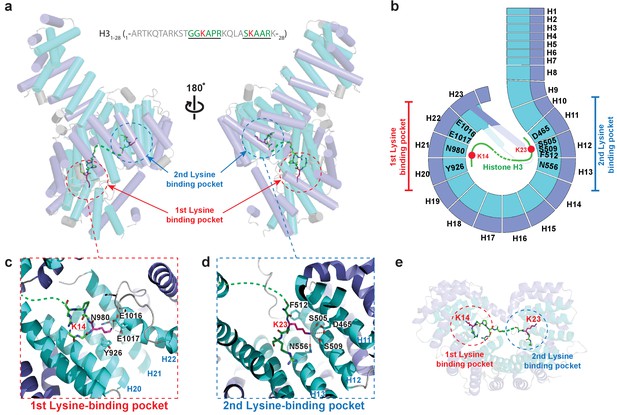
Crystal structure of Kl Kap123 in complex with H31–28-NLS.
(a) Crystal structure of full-length Kl Kap123 in the presence of H31–28-NLS (green stick model, 1-ARTKQTARKSTGGKAPRKQLASKAARK-28) with two lateral views (180° rotation). The two lysine-binding pockets located at the inner curvature of Kap123 are marked with red (first lysine-binding pocket) and blue (second lysine-binding pocket) dashed circles. Residues 12–17 and 21–26 of H31–28-NLS (chain B) are ordered and visible in the structure. The two lysine residues (H3 K14 and K23) that bind to these lysine-binding pockets of Kap123 are colored red. (b) Schematic view of Kl Kap123 in complex with H31–28-NLS. The residues and HEAT repeats that participate in organizing two lysine-binding pockets are described. (c) The first lysine-binding pocket of Kl Kap123. K14 of H31–28-NLS forms hydrophobic (Y926) and electrostatic/hydrogen bond (N980, E1016, and E1017) interactions with Kap123 through repeats 20–22. (d) The second lysine-binding pocket of Kl Kap123. K23 of H31–28-NLS makes hydrophobic (Y512) and electrostatic/hydrogen bond (D465, S505, S509, and N556) interactions with Kap123 through repeats 11–13. (e) Top view of Kl Kap123 in complex with H31–28-NLS. Two lysine-binding sites are distally located and the middle region of H31–28-NLS does not make any specific contacts with Kap123.
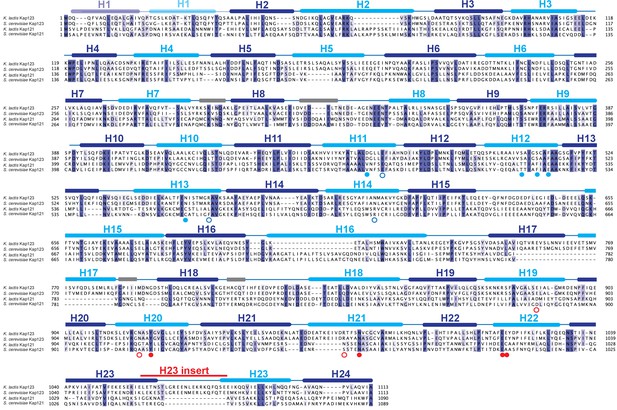
Multiple sequence alignment of Kap121 and Kap123 in budding yeast.
Primary sequences of K. lactis Kap123, S. cerevisiae Kap123, K. lactis Kap121 and S.cerevisiae Kap121 were aligned using Clustal Omega (Adams et al., 2010). The illustration for the alignment was generated with Jalview using default settings (Waterhouse et al., 2009). Highly conserved residues are shown in purple. The secondary structure identified from the K. lactis Kap123 crystal structure is shown above the sequence with the corresponding HEAT repeats with inner (cyan) and outer (blue) helices. Linker regions are colored in grey. The H23 insert region (Figure 1—figure supplement 1), a unique feature of K. lactis Kap123, is also indicated above the sequence. K. lactis Kap123 residues that form the first lysine-binding pocket (solid red circle) and the second lysine-binding pocket (solid blue circle) in the Kap123-H31-28-NLS complex are marked below the sequence accordingly. Residues involved in the H3-NLS peptide backbone interaction near the first lysine-binding pocket (open red circle) and the second lysine-binding pocket (open blue circle) in the Kap123-H31-28-NLS complex are also indicated below the sequence.
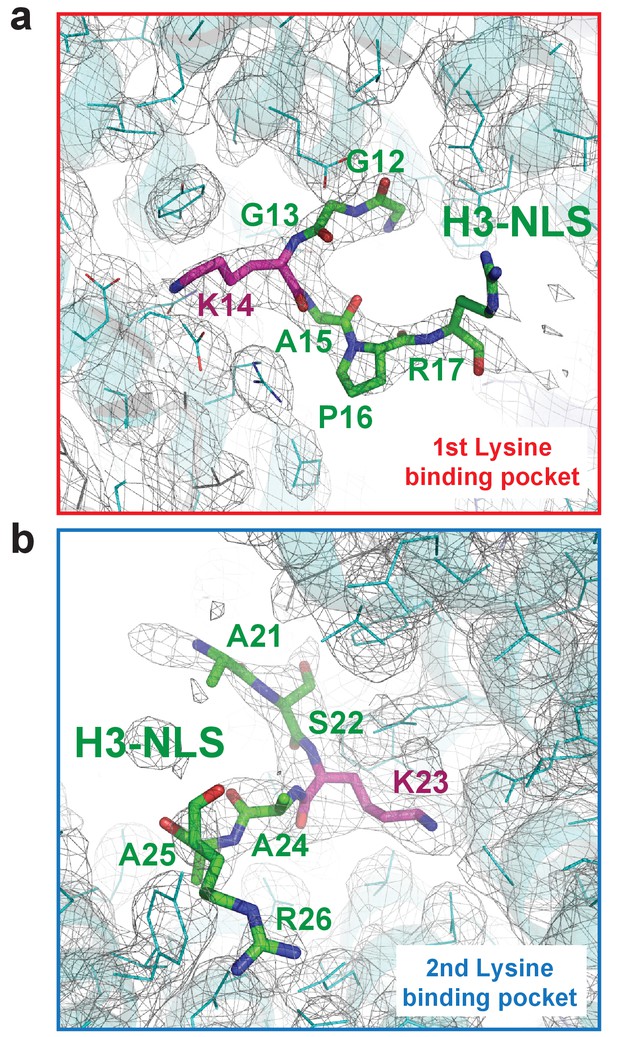
The Composite omit map (1.0 σ contour level) of the Kap123-H31-28-NLS complex.
The composite omit map of the first lysine-binding pocket (a) and the second lysine-binding pocket (b) of Kap123. The omit map was calculated in the absence of H3 peptide using Program Composite omit map in the PHENIX package (Sievers et al., 2011).
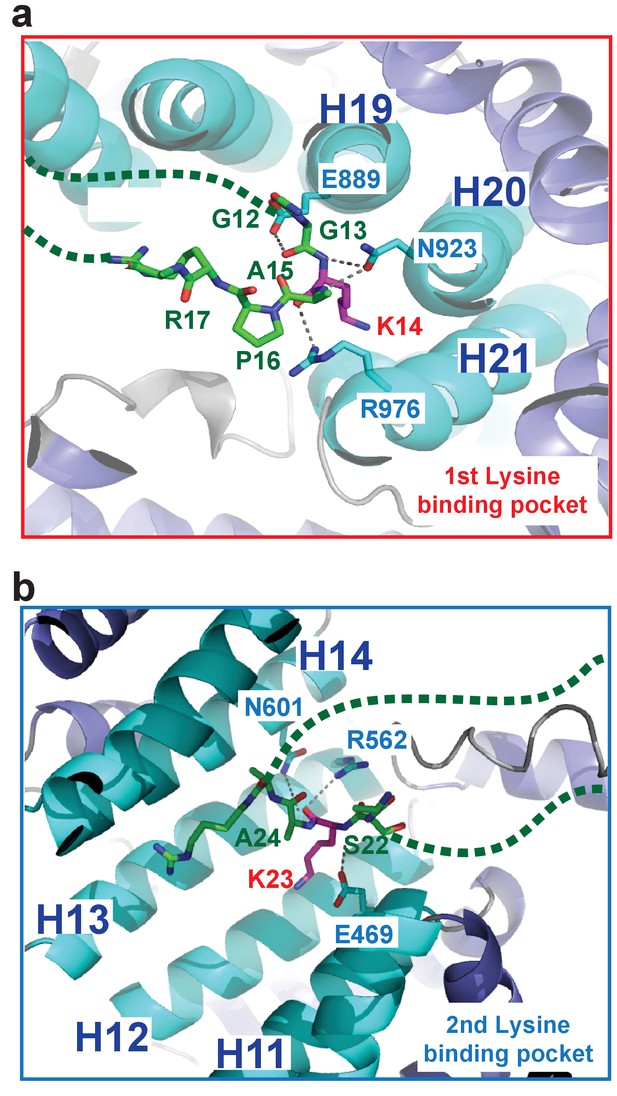
H31–28-NLS peptide backbone interactions in the Kap123-H31–28-NLS complex.
(a) H31–28-NLS peptide backbone interactions near the first lysine-binding pocket. Kap123 residues near the first lysine-binding pocket (repeats 19–21; E889, N923, and R976) form multiple hydrogen bond interactions with the peptide backbone of H31–28-NLS (stick model, green). (b) H31–28-NLS peptide backbone interactions near the second lysine-binding pocket. Kap123 residues near the second lysine-binding pocket (repeats 11–14; E469, R562, and N601) form multiple hydrogen bond interactions with the peptide backbone of H31–28-NLS (stick model, green). H3 K14 and K23 associate with the first and second lysine-binding pocket, respectively (red). Unstructured regions of H31–28-NLS are shown as green dashed lines.
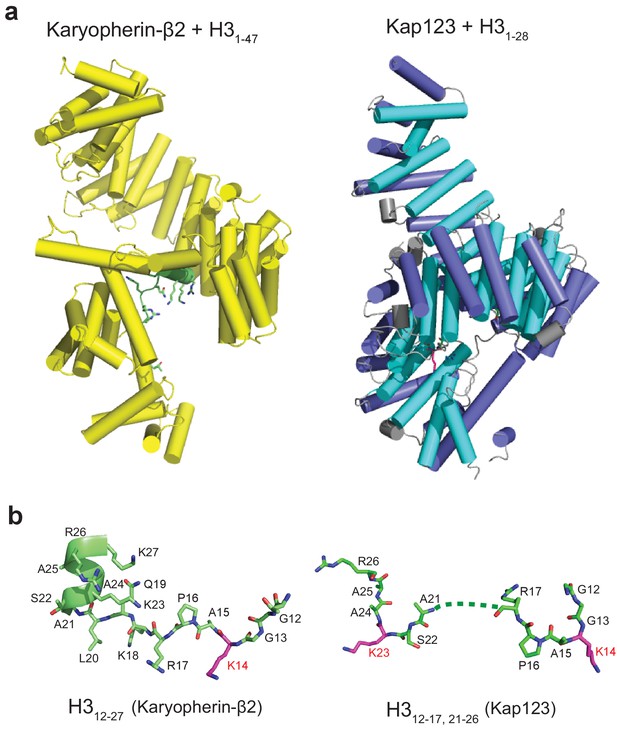
Structural comparison of Kap123-H31-28-NLS and Kapβ2-H31-47-NLS complexes.
(a) The side-views of both Kapβ2-H31-47-NLS (yellow, PDB ID: 5J3V) (Soniat and Chook, 2016) and Kap123-H31-28-NLS complexes. H3-NLS peptides in both structures are colored green with stick and cartoon model. (b) The conformation of H3-NLS peptides within the Kapβ2-H31-47-NLS and Kap123-H31-28-NLS structures. Residues 12–27 and 12-18/22-25 of histone H3 are only visible in Kapβ2-H31-47-NLS and Kap123-H31-28-NLS, respectively. Key lysine residues participating in Kap association (K14 in Kapβ2-H31-47-NLS and K14/K23 in Kap123-H31-28-NLS) are colored red. Residues 19–21 of H3-NLS in Kap123-H31-28-NLS are disordered and displayed as dashed line.
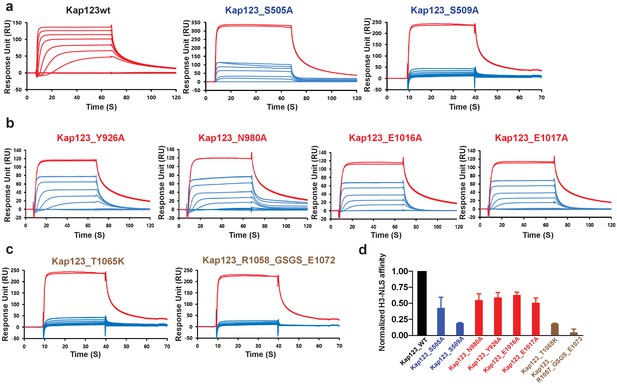
Surface plasmon resonance analysis of wild-type and mutants Kl Kap123 in the presence of H31–35-NLS.
The biacore diagrams of Kl Kap123 wild-type and mutants in the presence of H31–35-NLS peptides are listed. (a) Streptavidin was immobilized at 400 resonance units (RU) and the C-terminal biotinylated histone H3 peptide (residues 1–35) was captured at 150 RU on a CM5 chip. The wild-type Kap123 was injected with different concentrations (0, 1, 5, 10, 25, 50, 100, 250, and 500 nM) and sensorgrams are colored red. To monitor the affinity of Kap123 mutants at the second lysine-binding pocket (S505A and S509A), the C-terminal biotinylated histone H3 peptide was captured (300 RU) on a streptavidin-immobilized CM5 chip (400 RU). Various concentrations of Kap123_S505A (0, 65, 125, 250, 500, 1000, and 2000 nM) and Kap123_S509A (0, 32, 65, 125, 250, 500, 1000, and 2000 nM) were injected; their sensorgrams are shown in blue. (b) Affinity measurement of first lysine-binding pocket mutants of Kap123 (Y926A, N980A, E1016A, and E1017A). Streptavidin (400 RU) and C-terminal biotinylated histone H3 peptide (150 RU) were immobilized on a CM5 chip. The mutant proteins were injected with various concentrations (0, 1, 5, 10, 25, 50, 100, 250, and 500 nM) and sensorgrams are shown in blue. (c) Affinity measurement of Kap123 mutants in the extended helix of the repeat 23. Either the point mutation (T1065K) or the deletion mutant (1058-GSGS-1072; residues 1059–1071 are replaced by the GSGS linker) of Kap123 was injected with various concentrations (0, 32, 65, 125, 250, 500, 1000, and 2000 nM for T1065K and 0, 1, 5, 10, 25, 50, 100, 250, and 500 nM for 1058-GSGS-1072). Red-colored sensorgrams are positive controls of wild-type Kap123, which we injected at 2000 nM wild-type Kap123 before and after sample injection (a–c). (d) Normalized affinity of wild-type and mutant Kap123s toward H3-NLS is shown in the bar graph. The relative affinity of wild-type and mutant Kap123s measured at five different concentrations is used to generate error bars.
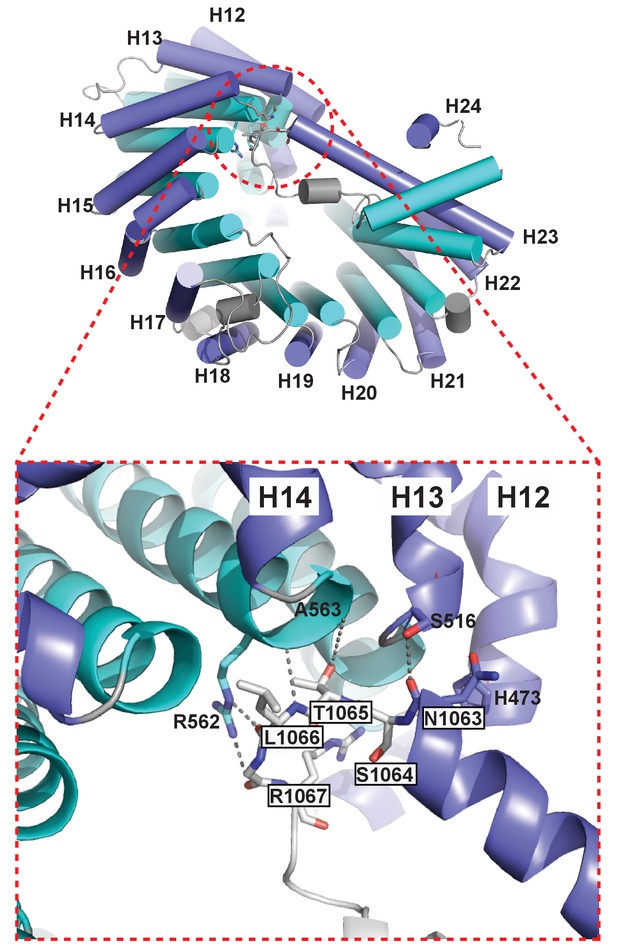
The extended helix of Kl Kap123 repeat 23 makes intramolecular interactions with repeats 12–14.
Residues from the extended helix of the repeat 23 (residue name white and boxed) generate multiple hydrogen bond and electrostatic interactions with residues at the ridge of repeats 12–14 (residue name without box).
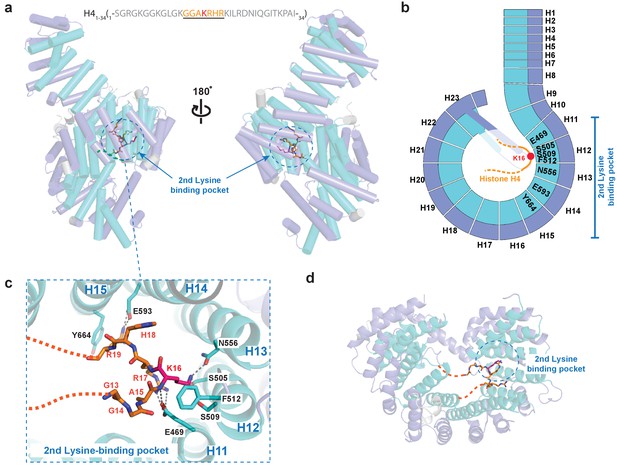
Crystal structure of Kl Kap123 in complex with H41–34-NLS.
(a) The crystal structure of full-length Kl Kap123 in the presence of H41–34-NLS (orange stick model, 1-SGRGKGGKGLGKGGAKRHRKILRDNIQGITKPAI-34) with two lateral views (180° rotation). The second lysine-binding pocket located at the inner curvature of Kap123 is marked as a blue dashed circle. Residues 13–20 of H41–34-NLS are visible in the structure. H4 K16, which binds to the second lysine-binding pocket, is colored red. (b) Schematic view of Kl Kap123 in complex with H41–34-NLS. Residues of repeats 11–15 that participate in H41–34-NLS recognition are indicated. (c) The second lysine-binding pocket of Kl Kap123 in H41–34-NLS recognition. K16 of H41–34-NLS forms hydrophobic (F512) and electrostatic/hydrogen bond (S505, S509, and N556) interactions with Kap123 through repeats 12–13. R17 and R19 of H41–34-NLS form additional electrostatic and hydrophobic contacts with E469 (repeat 11) and E593 (repeat 14)/Y664 (repeat 15) of Kap123, respectively. (d) Top view of Kl Kap123 in complex with H41–34-NLS. Only the second lysine-binding pocket is occupied by K16 of H41–34-NLS.
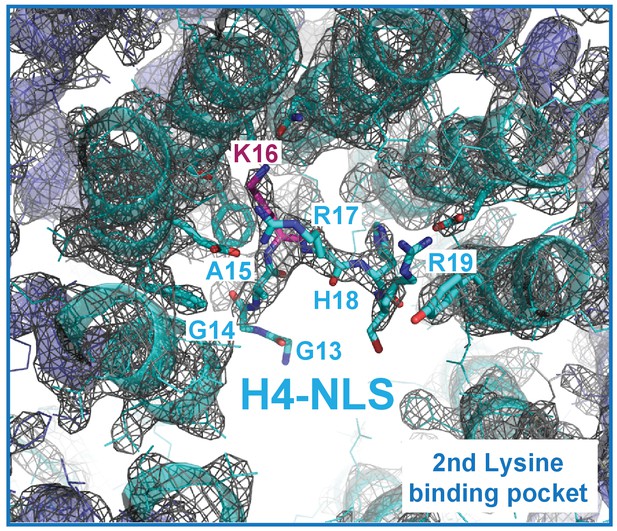
The composite omit map (1.0 σ level) of the Kap123-H41–34-NLS complex.
The composite omit map of the Kap123 second lysine-binding pocket was calculated in the absence of H4 peptide using Program Composite omit map in the PHENIX package (Sievers et al., 2011).
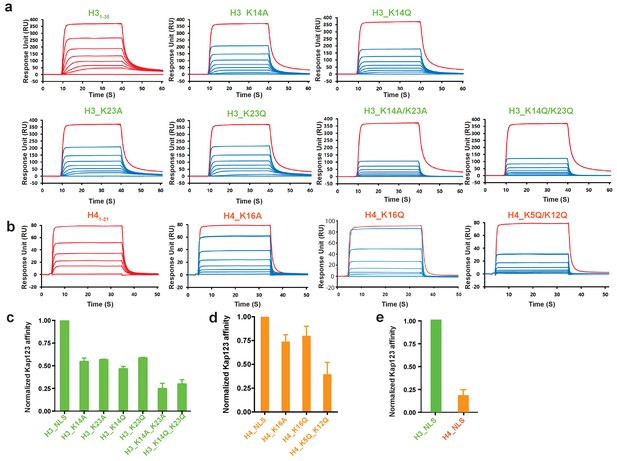
Surface plasmon resonance analysis of wild-type Kl Kap123 in the presence of wild-type or mutants of H31–35-/H41–21-NLSs.
The biacore diagrams (a) and the normalized bar graph (c) of wild-type Kap123 in the presence of wild-type or mutant H31-35-NLS. The wild-type Kap123 was immobilized on a CM5 chip with 7500 RU and the wild-type or mutant H31–35-NLS peptides (H3_K14A, H3_K14Q, H3_K23A, H3_K23Q, or H3_K14Q/K23Q) were injected at different concentrations (0, 0.07, 0.15, 0.32, 0.65, 1.25, 2.5, and 5 μM). The biacore diagrams (b) and the bar graph (d) of wild-type Kap123 in the presence of wild-type or mutant H41–21-NLS. The wild-type Kap123 was immobilized on a CM5 chip with 7116 RU and the wild-type or mutant H41–21-NLS peptides (H4_K16A or H4_K5Q/K12Q) were injected at different concentrations (0, 0.32, 0.62, 1.25, 2.5, 5, and 10 μM). The H4K16Q peptide was used at different concentrations (0, 0.7, 1.5, 3, 6, 12, and 25 uM). See details in Materials and method section. Red-colored sensorgrams represent positive controls of wild-type Kap123, which were injected 2000 nM wild-type Kap123 before and after sample injection (a–b). The relative binding between wild-type Kap123 and mutant peptides was measured at five different concentrations for generating error bars (c–e). (e) The normalized affinity bar graph of H3-/H4-NLS toward wild-type Kap123 was derived from (a) and (b).
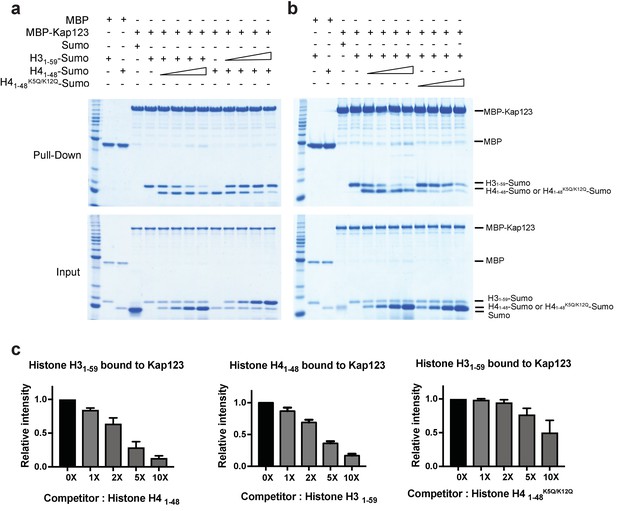
Competition assay of H3- and H4-NLSs (wild-type H4 and mutant H4K5Q/K12Q) toward Kap123 association.
Competition of H31–59- and H41–48-Sumo (a) as well as H31–59- and H41–48 K5Q/K12Q-Sumo (b) toward Kap123 binding was monitored by amylose affinity pull-downs using MBP-tagged Kap123. A fixed amount of the pre-assembled complex of MBP-tagged Kap123 and H41–48-Sumo was challenged by an increasing amount of H31–59-Sumo. The same competition assay was done by using pre-incubated MBP-tagged Kap123-H31–59-Sumo and gradually increasing the amount of H41–48-Sumo either with wild-type (a) or mutant (b), K5Q/K12Q) H4 sequence. (c–d) The normalized bar graphs of remaining H31–59- and H41–48-NLS-Sumo after challenged with increased amounts of H41–48- and H31–59-NLS-Sumo (0x, 1x, 2x, 5x, and 10x) toward Kap123, respectively. Either H41–48-NLS-Sumo (c) or H41–48K5Q/K12Q-NLS-Sumo (d) was used for the H31–59-NLS-Sumo competition. Three independent competition assays were carried out and the band intensity was measured to calculate the error bar.
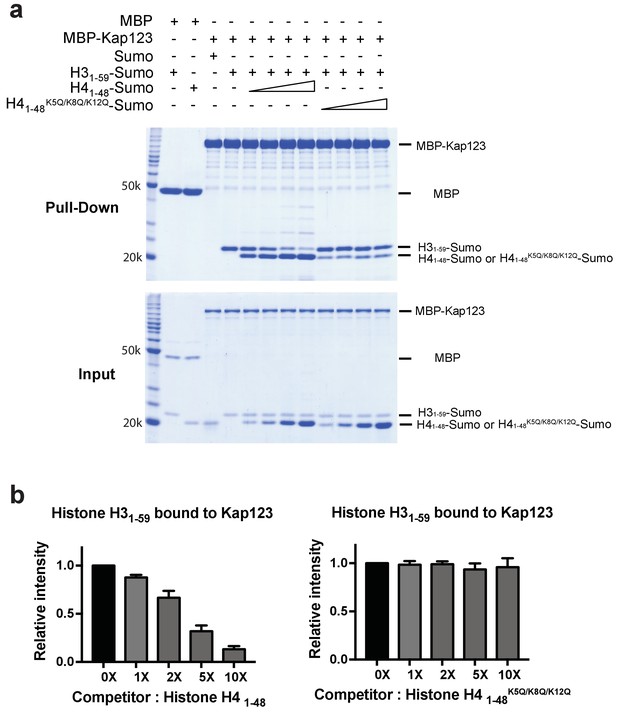
Competition assay of H3- and H4-NLSs (wild-type H4 and mutant H4K5Q/K8Q/K12Q) toward Kap123 association.
(a) Competition of H31–59-Sumo using either H41–48-Sumo or H41–48 K5Q/K8Q/K12Q-Sumo toward Kap123 binding was monitored by amylose affinity pull-downs using MBP-tagged Kap123. A fixed amount of the pre-assembled complex of MBP-tagged Kap123 and H31–59-Sumo was challenged by an increasing amount of H41–48- or H41–48 K5Q/K8Q/K12Q-Sumo. (b) The normalized bar graphs of remaining H31–59-Sumo after challenged with increased amounts (0x, 1x, 2x, 5x, and 10x) of H41–48-Sumo or H41–48K5Q/K8Q/K12Q-Sumo. Three independent competition assays were carried out and the band intensity was measured to calculate the error bar.
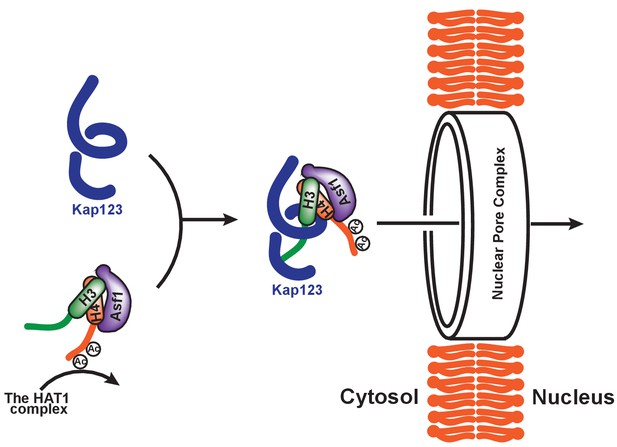
Proposed model of Kap123-dependent nuclear translocation of the H3:H4/Asf1 complex.
Schematic model of the potential role of histone H4 diacetylation during nuclear import. Newly synthesized histones H3 and H4 are associated and immediately protected by its specific chaperone, Asf1. The HAT1 complex subsequently acetylates K5 and K12 of histone H4 as a part of the H3:H4/Asf1 complex. Diacetylation of the H4-NLS, whose affinity toward Kap123 is already fivefold weaker than H3-NLS, further destabilizes the Kap123-histone H4 interaction. Therefore, Kap123 preferentially associates with the H3-NLS and allows for histone H3-dependent Kap123 association during nuclear translocation. It should be noted that there are several histone H3 variants available in eukaryotes but there is only one known histone H4 protein, which can be commonly shared by each histone H3 variant.
Additional files
-
Supplementary file 1
Data collection and refinement statistics.
- https://doi.org/10.7554/eLife.30244.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30244.018





