Dynamic clustering of dynamin-amphiphysin helices regulates membrane constriction and fission coupled with GTP hydrolysis
Figures
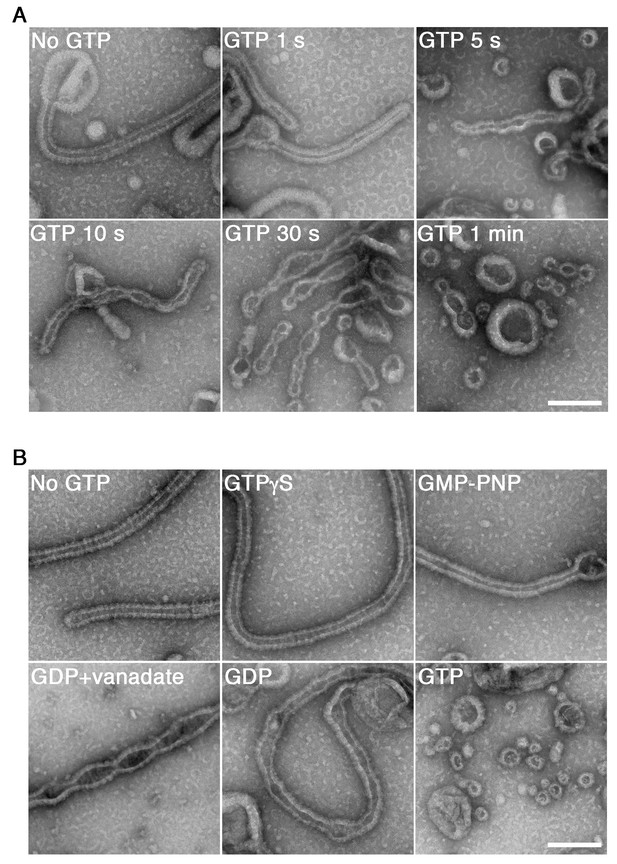
GTP hydrolysis is required and sufficient for membrane constriction by dynamin-amphiphysin helical complexes.
(A) Electron micrographs of lipid tubules induced by dynamin-amphiphysin helical complex before GTP addition (No GTP) and at different time points after addition of 1 mM GTP (GTP 1 s, GTP 5 s, GTP 10 s, GTP 30 s and GTP 1 min). More than thirty samples from three individual experiments were examined and representative images are shown. Scale bar is 200 nm. (B) Electron micrographs of lipid tubules induced by dynamin-amphiphysin helical complex without guanine nucleotide (No GTP) or with a transition states analogue of GTPase reaction, by adding 1 mM each of slowly hydrolyzable GTP analogue (GTPγS), nonhydrolyzable GTP analogue (GMP-PNP), GDP combined with vanadate (GDP + vanadate), GDP (GDP), or GTP (GTP) for 10 min. More than thirty samples from three individual experiments were examined and representative images are shown. Scale bar is 200 nm.
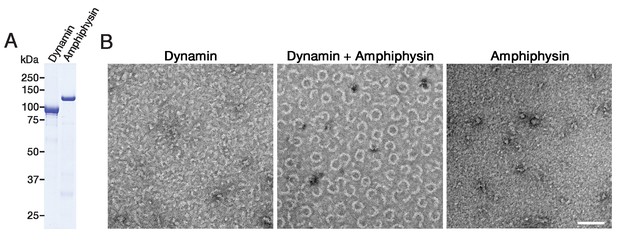
Purified dynamin and amphiphysin forms ring-shaped complexes.
(A) SDS-PAGE of purified dynamin (Dynamin) and amphiphysin (Amphiphysin). (B) Negative staining electron micrographs of dynamin alone (Dynamin), dynamin and amphiphysin (Dynamin + Amphiphysin) and amphiphysin alone (Amphiphysin) incubated in cytosolic buffer with physiological salt concentration and PH (see Materials and methods). Scale bar is 100 nm.
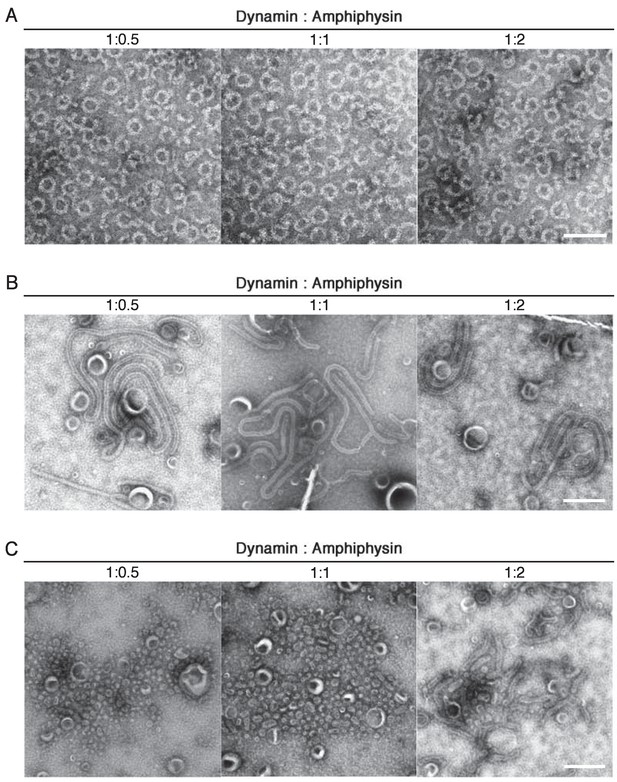
Stoichiometry dependency of dynamin and amphiphysin in ring complex formation, liposome tubulation and membrane fission.
Dynamin (2.3 μM) were mixed with 1.15 μM, 2.3 μM and 4.6 μM of amphiphsyin (1:0.5, 1:1 and 1:2 in molar ratio) and formation of the ring-shaped complexes (A), tubulation of liposomes (B) and membrane fission (C) were analyzed by EM. Scale bars are 100 nm and 500 nm for (A) and (B), (C), respectively.
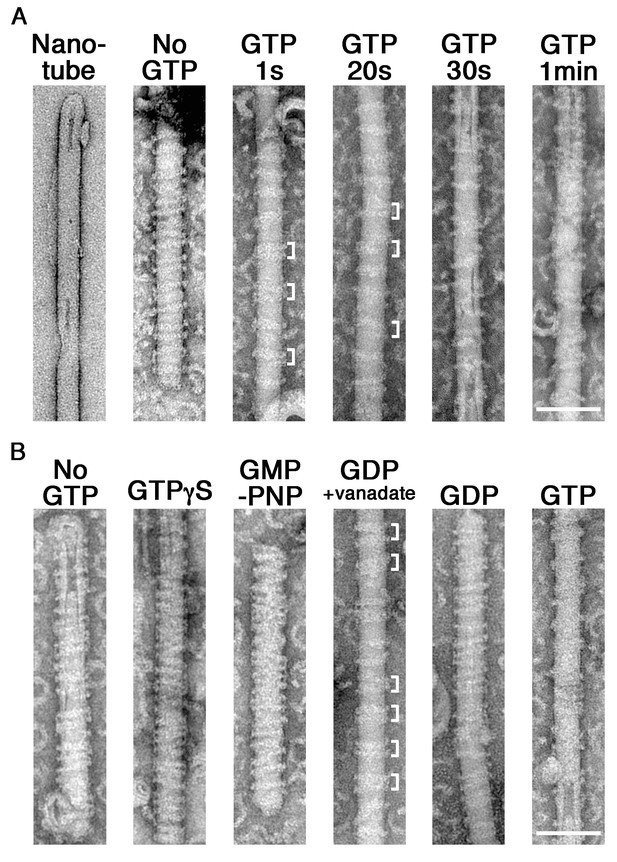
GTP hydrolysis induces clustering of dynamin-amphiphysin complexes on lipid nanotubes.
(A) Electron micrographs of a lipid nanotube (Nanotube) and those with dynamin-amphiphysin complexes before GTP addition (No GTP) and at different time points after GTP addition (GTP 1 s, GTP 20 s, GTP 30 s, and GTP 1 min). Clusters of dynamin-amphiphysin helical complexes are indicated (white brackets). More than thirty samples from three individual experiments were examined and representative images are shown. Scale bar is 100 nm. (B) Electron micrographs of lipid nanotubes after addition of dynamin-amphiphysin complexes without guanine nucleotide (No GTP) or with a transition states analogue of GTPase reaction, by adding 1 mM each of slowly hydrolysable GTP analogue (GTPγS), nonhydrolyzable GTP analogue (GMP-PNP), GDP combined with vanadate (GDP + vanadate), GDP (GDP) and GTP (GTP) for 10 min. More than 30 samples from three individual experiments were examined and representative images are shown. Clusters of dynamin-amphiphysin helical complexes are indicated (white brackets). The average pitch of helices in the clusters is 15.00 ± 2.2 nm (mean pitch ± s.e.m., n = 63 from 7 nanotubes) in GDP + vanadate, while the average pitch of the helical complexes is 20.0 ± 0.5 nm (mean pitch ± s.e.m., n=81 from 9 nanotubes) in No GTP control. Scale bar is 100 nm.
-
Figure 2—source data 1
Measuring pitch size of dynamin-amphiphysin helices on the lipid nanotube without GTP and in the presence of GDP and vanadate in panel B.
- https://doi.org/10.7554/eLife.30246.008
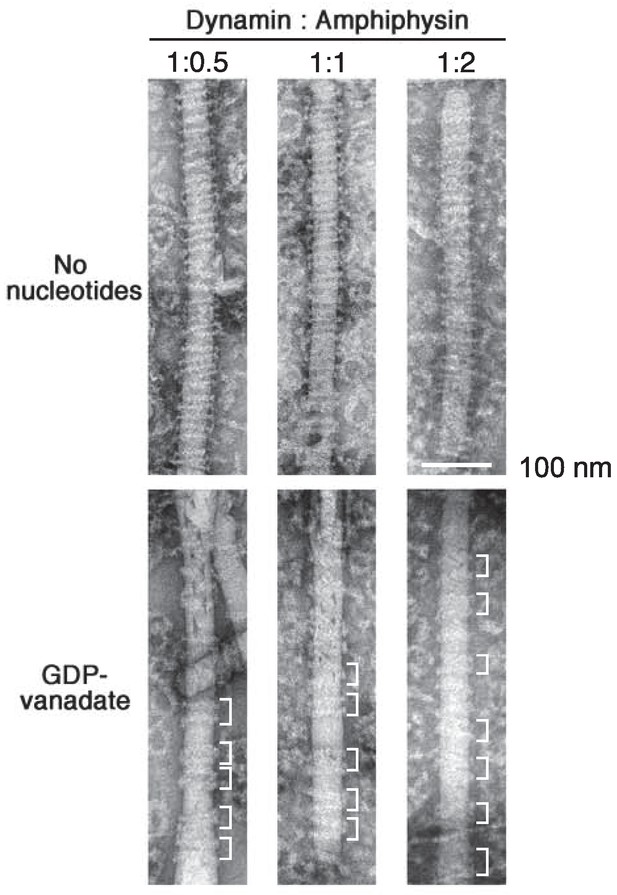
Stoichiometry dependency of dynamin and amphiphysin in cluster formation
Dynamin (2.3 μM) were mixed with 1.15 μM, 2.3 μM and 4.6 μM of amphiphsyin (1:0.5, 1:1 and 1:2 in molar ratio) and their structure on lipid nanotubes in the absence of nucleotides (no nucleotides) and in the presence of 1 mM GDP and vanadate (GDP-vanadate) were analyzed by EM. Clusters formed by a few helical turns of dynamin-amphiphysin complexes are indicated (white brackets). Scale bar is 100 nm.
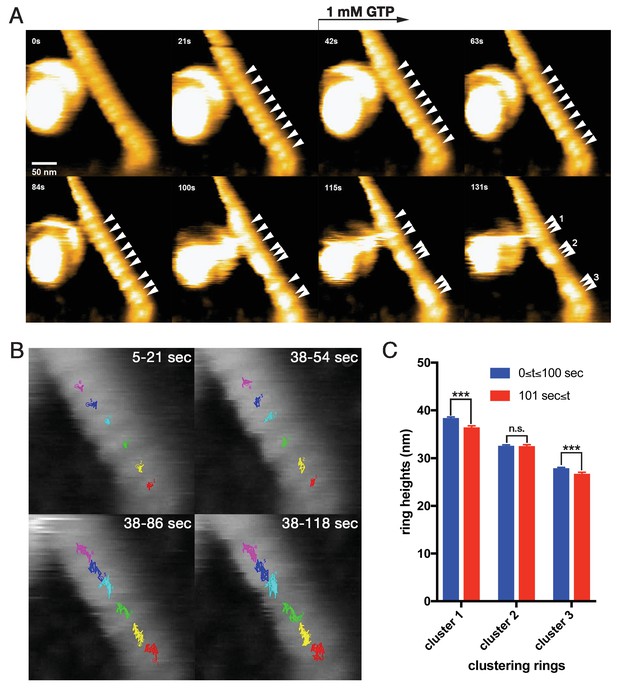
Dynamic clustering of dynamin-amphiphysin helices during GTP hydrolysis.
(A) HS-AFM images captured at 1 frame/s of dynamin-amphiphysin helical complexes on membrane tubules before (0 s and 21 s) and after GTP addition at different time points (42 s, 63 s, 84 s, 100 s, 115 s and 131 s). Dynamin-amphiphysin helices (arrowheads) are assembled into three distinct clusters (1, 2 and 3 at 131 s). The pitch of dynamin-amphiphysin helices on the lipid tubule was 22.0 ± 0.7 nm (mean pitch ± s.e.m., n = 36 from 3 time points) before GTP addition and 15.7 ± 0.3 nm (mean pitch ± s.e.m., n = 36 from 9 time points) after GTP addition. (B) Particle tracking of dynamin-amphiphysin helices before (5–21 s) and after addition of 1 mM GTP (38–54 s, 38–86 s and 38–118 s) from Videos 4 and 5, respectively. Particle tracking of the complexes in the cluster 2 (light blue, dark blue and magenta) and cluster 3 (red, yellow and green) are shown. (C) Dynamin-amphiphysin helices tend to constrict during clustering. Average heights before (0 ≤ t ≤ 100 s) and after clustering (101 s ≤ t) are 38.4 ± 0.2 nm and 36.5 ± 0.2 nm for cluster 1, 32.6 ± 0.2 nm and 32.6 ± 0.2 nm for cluster 2, 27.9 ± 0.1 nm and 26.8 ± 0.2 nm for cluster 3, respectively. The heights were measured from the substrate surface. The marks *** indicate p<0.001 and n.s. is not significant, respectively.
-
Figure 3—source data 1
Measuring pitch size of dynamin-amphiphysin helices on the lipid tubule before and after cluster formation.
Pitch sizes of dynamin-amphiphysin helices before cluster formation (0, 3 and 6 s) and after cluster formation (111–119 s) were measured based on the Video 3 for panel A.
- https://doi.org/10.7554/eLife.30246.012
-
Figure 3—source data 2
Measuring heights of dynamin-amphiphysin helices on the lipid tubule before and after cluster formation.
Average heights of dynamin-amphiphysin helices composing clusters 1, 2 and 3 before cluster formation (0 ≤ t ≤ 100 s) and after cluster formation (101 ≤ t s) were calculated based on Figure 3—figure Supplement 2—source data 1 for panel C.
- https://doi.org/10.7554/eLife.30246.013
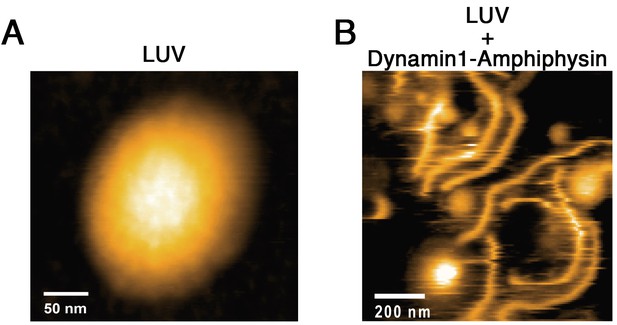
HS-AFM imaging of LUV and its tubulation by dynamin-amphiphysin complex.
(A) HS-AFM images of a LUV. Scale bar is 50 nm. (B) Lipid tubules induced from LUVs in the presence of dynamin-amphiphysin complexes. Scale bar is 200 nm.
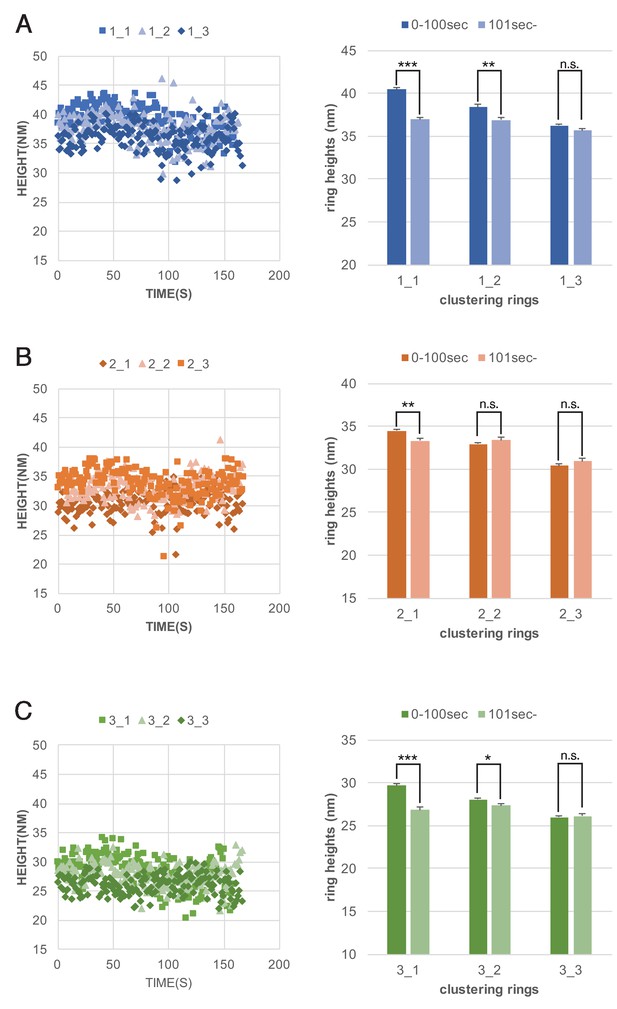
Dynaimin-amphiphysin helices constrict during cluster formation.
Maximum heights of each helices plotted as a function of time (left panels) and their average helix heights before (0–100 s) and after clustering (101sec-) are shown for clusters 1, 2 and 3 (A, B and C, respectively). ***: p<0.001, **: p<0.01, *: p<0.1, n.s.: not significant.
-
Figure 3—figure supplement 2—source data 1
Measuring heights of dynamin-amphiphysin helices on the lipid tubule during cluster formation.
Height changes of each helices composing clusters 1, 2 and 3 were measured based on the Video 3. Average heights of each dynamin-amphiphysin helices composing clusters 1, 2 and 3 before cluster formation (0 ≤ t ≤ 100 s) and after cluster formation (101 ≤ t s) were also calculated.
- https://doi.org/10.7554/eLife.30246.014
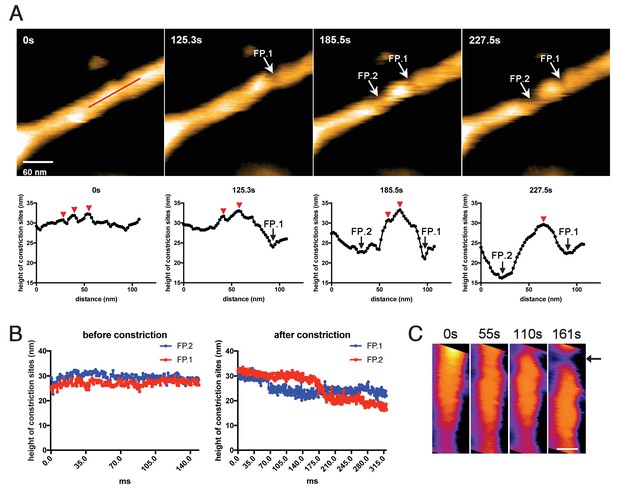
Membrane fission occurs at the protein-uncoated regions flanking dynamin-amphiphysin clusters.
(A) Clips of HS-AFM images captured at 0.42 frames/s showing membrane fission by dynamin-amphiphysin complexes (0 s, 125.3 s, 185.5 s and 227.5 s) in Video 6. Membrane fission occurred at flanking regions of a dynamin-amphiphysin cluster. Corresponding height profiles along the red line (shown in the 0 s image) passing through the two fission points (arrows marked with FP.1 and FP.2) are shown below, together with clustered dynamin-amphiphysin helical complexes (red arrowheads). (B) Height profiles at fission points (FP.1 and FP.2) over time before (Video 7) and after constriction (Video 6). The heights of the lipid tubules from the substrate surface were measured at the fission points. (C) Clips of HS-AFM images showing clustering dynamin-amphiphysin complexes and membrane constriction at flanking regions of the cluster (arrow). HS-AFM images are shown in pseudo color. Scale bar is 40 nm.
-
Figure 4—source data 1
Measuring height changes of lipid tubules during constriction and fission by dynamin-amphiphysin helices.
- https://doi.org/10.7554/eLife.30246.021
-
Figure 4—source data 2
Measuring heights of fission points (FP.1 and FP.2) over time before (Video 7) and after GTP addition (Video 6) for panel B.
- https://doi.org/10.7554/eLife.30246.022
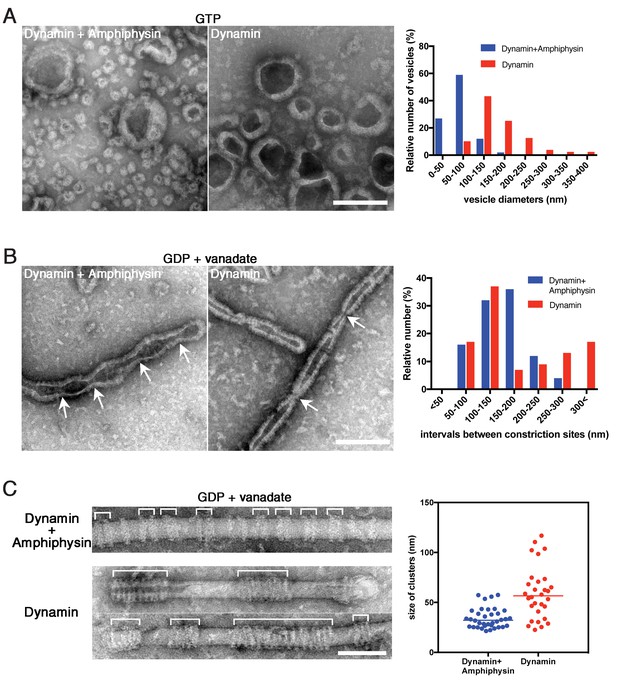
Amphiphysin contributes to generation of uniformly-sized vesicles by controlling dynamin-amphiphysin clusters.
(A) Representative EM images of membrane vesicles generated by dynamin-amphiphysin complexes (Dynamin + Amphiphysin) or dynamin alone (Dynamin) after addition of GTP. Size distribution of generated vesicles are shown in the right panel. The average sizes of vesicles were 70.0 ± 0.6 nm (mean diameter ± s.e.m., n > 30, N = 3) for dynamin-amphiphysin complexes and 204.6 ± 1.1 nm (mean diameter ± s.e.m., n > 45, N = 3) for dynamin alone. Scale bar is 200 nm. (B) Representative EM images of membrane constriction induced by dynamin-amphiphysin complexes (Dynamin + Amphiphysin) and dynamin alone (Dynamin) in the presence of GDP and vanadate. Distribution of intervals between constriction sites (arrows) are quantified in the right panel. The average intervals of constriction sites induced are 150.3 ± 9.8 nm (mean intervals ± s.e.m., n = 25 from 7 tubes) by dynamin-amphiphysin complexes and 193.5 ± 15.8 nm (mean intervals ± s.e.m., n = 46 from 15 tubes) by dynamin alone. Scale bar is 200 nm. (C) Clustering of dynamin-amphiphysin complexes (Dynamin + Amphiphysin) and dynamin alone (Dynamin) on lipid nanotubes in the presence of GDP and vanadate. Clusters of dynamin-amphiphysin helices are indicated (white brackets). Distribution of cluster size were shown as scattered plot in the right panel. Average size of the clusters formed by dynamin-amphiphysin complexes and dynamin alone are 34.2 ± 1.7 nm (mean cluster size ± s.e.m., n = 36 from 7 tubes) and 59.3 ± 4.7 nm (mean cluster size ± s.e.m., n = 30 from 5 tubes) respectively. Scale bar is 100 nm.
-
Figure 5—source data 1
Measuring diameters of vesicles generated by dynamin-amphiphysin complexes or dynamin alone after GTP addition for panel A.
- https://doi.org/10.7554/eLife.30246.027
-
Figure 5—source data 2
Measuring distances between membrane constriction sites induced by dynamin-amphiphysin complexes and dynamin alone in the presence of GDP and vanadate for panel B.
- https://doi.org/10.7554/eLife.30246.028
-
Figure 5—source data 3
Measuring size of clusters formed by dynamin-amphiphysin complexes and dynamin alone in the presence of GDP and vanadate for panel C.
- https://doi.org/10.7554/eLife.30246.029
Videos
HS-AFM imaging of a LUV.
https://doi.org/10.7554/eLife.30246.015HS-AFM imaging of a lipid tubules induced from LUVs by dynamin-amphiphysin complexes.
https://doi.org/10.7554/eLife.30246.016HS-AFM imaging of cluster formation by dynamin-amphiphysin helical complexes.
https://doi.org/10.7554/eLife.30246.017Particle tracking of dynamin-amphiphysin helical complexes before GTP addition (frames from 5 s to 21 s in Video 3).
https://doi.org/10.7554/eLife.30246.018Particle tracking of dynamin-amphiphysin helical complexes after GTP addition (frames from 38 s to 124 s in Video 3).
https://doi.org/10.7554/eLife.30246.019HS-AFM imaging of constriction and fission of lipid tubules by dynamin-amphiphysin complexes.
https://doi.org/10.7554/eLife.30246.023HS-AFM imaging of lipid tubules before constriction and fission by dynamin-amphiphysin complexes.
https://doi.org/10.7554/eLife.30246.024HS-AFM imaging of constriction and fission of lipid tubules by dynamin-amphiphysin complexes.
https://doi.org/10.7554/eLife.30246.025Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30246.030





