CRISPR/Cas9 and active genetics-based trans-species replacement of the endogenous Drosophila kni-L2 CRM reveals unexpected complexity
Figures
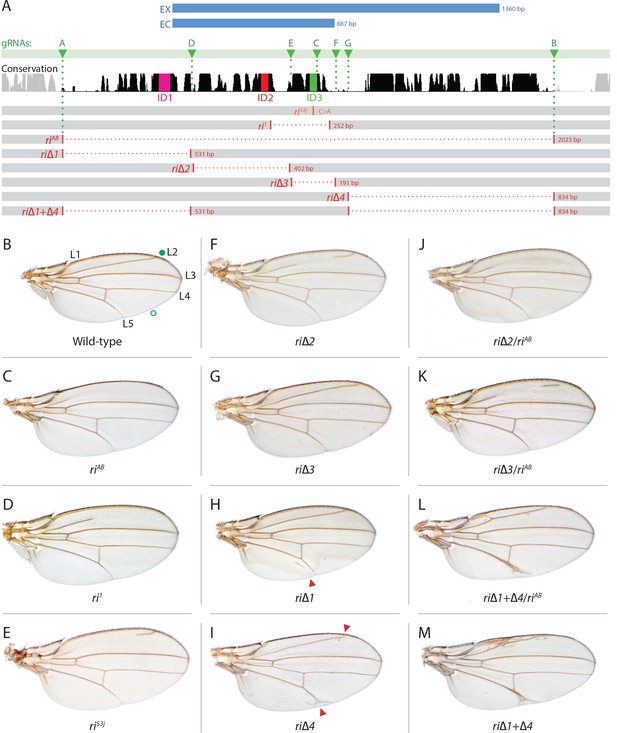
Fine scale in-locus deletion analysis of the D mel. kni L2-CRM.
(A) Map of kni L2-CRM and mutants. Diagram displays features of the cis-regulatory module (CRM) of the kni locus that restricts expression of the closely related kni and knrl genes in the L2 vein primordium. The black bar-scape indicates regions of sequence conservation, the green bar, arrows and dotted lines indicate the positions of guide RNAs (gRNAs, lettered A-G) used for dissection of this locus, the blue bars at the top indicate CRM-reporter gene fusions that have been previously analyzed (Lunde et al., 2003). The region present in EX and absent in EC were shown to contain sequences mediating repression of kni expression in peripheral regions of the wing. The ‘in-locus’ deletions generated in this study as well as known lesion (kniri53 and kniri1) are represented as gaps with a dotted red line along with details of the exact size of the deletion. These mutants were generated by injecting embryos expressing Cas9 with plasmids encoding the indicated gRNAs. (B–M) Adult wings of the following genotypes: (B) wild-type (WT) labeled with a filled or open green circle indicating anterior and posterior Sal borders, respectively, where Kni is expressed (see Figure 2A,M–O); C) homozygous AB deletion = kniriAB/kniriAB. (D) kniri1/kniri1; (E) kniri53j/kniri53j; (F) kniri∆2/kniri∆2; (G) kniri∆3/kniri∆3; (H) kniri∆1/kniri∆1. (I) kniri∆4/kniri∆4; (J) kniri∆2/kniriAB; (K) kniri∆3/kniriAB; (L) kniri∆1+∆4/kniriAB; (M) kniri∆1+∆4/kniri∆1+∆4.
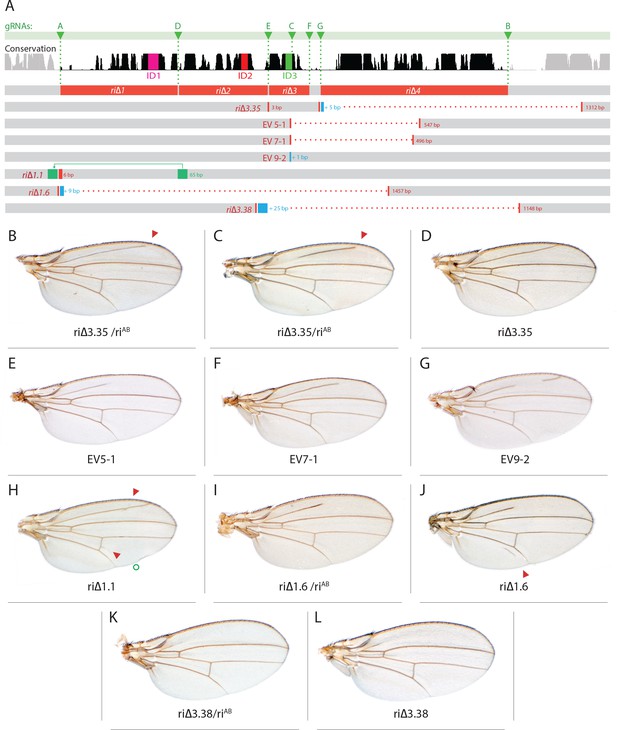
Wing phenotypes associated with fortuitously recovered CRISPR L2-CRM alleles.
(A) Map of L2-CRM showing locations and nature of mutations relative to those generated intentionally in Figure 1. Symbols as in Figure 1. Additional deleted sequences depicted by red boxes and added sequences by blue boxes. For the ri∆1.1 allele, a duplicated region that has been inserted upstream of its normal position is indicated by a green box and the accompanying arrow points to the novel insertion site. (B) kniri∆3.35/kniriAB, weak phenotype (just tip of L2 missing); (C) kniri∆3.35/kniriAB, strong phenotype; (D) kniri∆3.35/kniri∆3.35 homozygote has wild-type phenotype; (E) kniriEV5-1/kniriEV5-1; (F) kniriEV7-1/kniriEV7-1. (G) kniriEV9-2/kniriEV9-2; (H) kniri∆1.1/kniri∆1.1; (I) kniri∆1.6/kniriAB; (J) kniri∆1.6kniri∆1.6. (K) kniri∆3.38/kniriAB; (L) kniri∆3.38/kniri∆3.38.
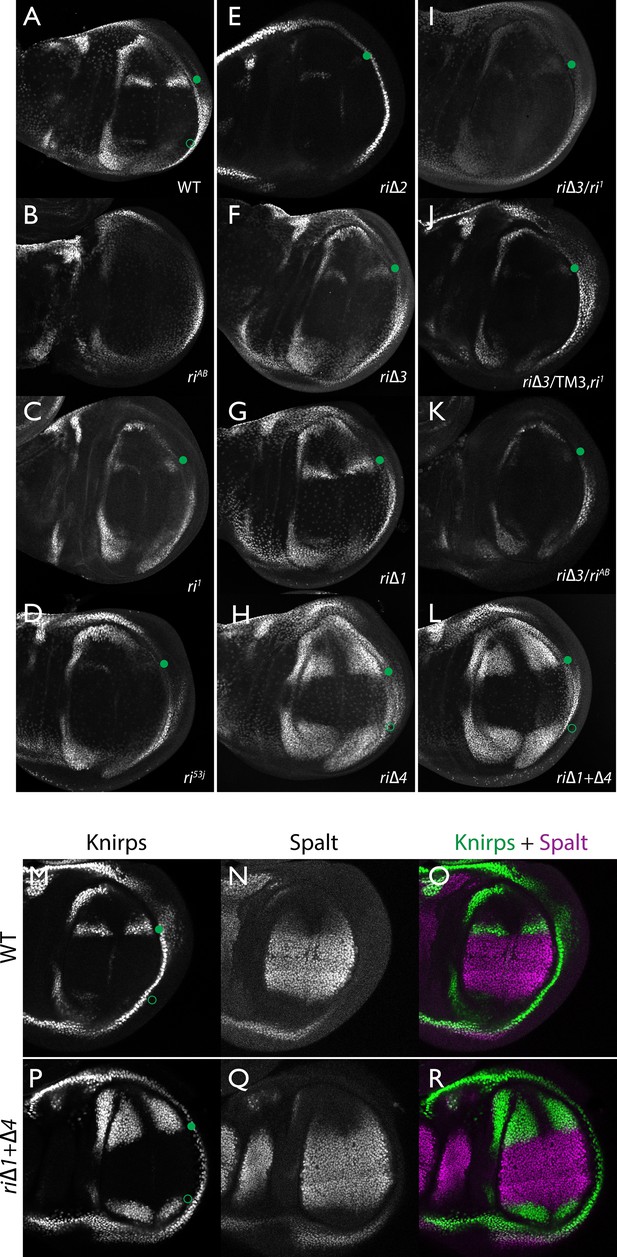
Endogenous Kni expression resulting from CRISPR generated in-locus CRM mutations.
Kni expression in late third instar wing imaginal discs was determined by staining with a polyclonal anti-Kni antibody alone (A–L) or in combination with an anti-Spalt antibody (M–R). Genotypes are specified by the labeling of each panel, marked with a filled green circle indicating location of anterior Kni expression and an open green circle indicating faint posterior expression along the Sal border. In panels (M–R), stains for Kni and Spalt are shown separately as black and white images and combined as green (Kni) and magenta (Sal) images.
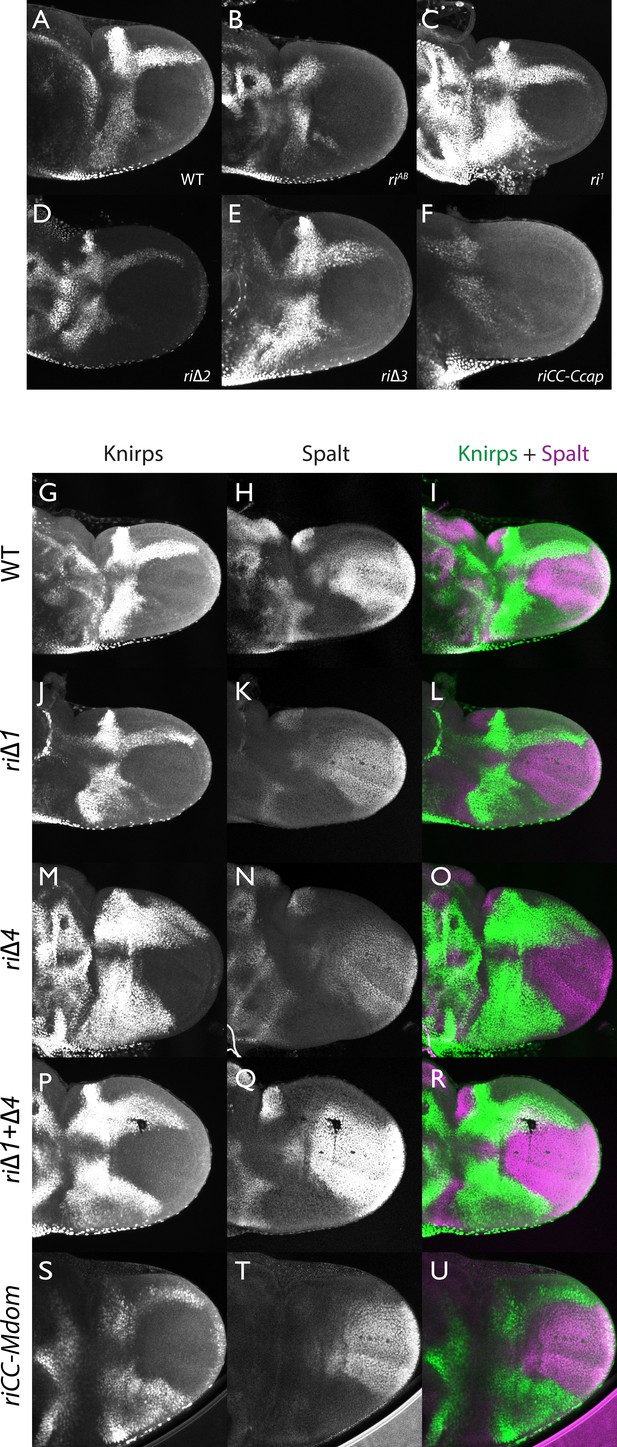
Knirps and Spalt expression in prepupal wings.
Expression of Kni and Sal expression in prepupal wings (4–6 hr after pupariation). (A–F): Kni single stains: (A) wild-type (WT); (B) kniriAB/kniriAB; (C) kniri1/kniri1; (D) kniri∆2/kniri∆2; (E) kniri∆3/kniri∆3; (F) kniriCC-Ccap>/kniriCC-Ccap. (G–U): Kni (green) + Sal (magenta) double stains: (G-I) wild-type; J–L) kniri∆1/kniri∆1. (M–O) kniri∆4/kniri∆4; P–R) kniri∆1+∆4/kniri∆1+∆4; (S–U) kniriCC-Mdom/kniriCC-Mdom. Note that the zone of ectopic anterior Kni expression is narrower in the kniri∆1+∆4 double mutant (P,R) than in the kniri∆4 single mutant (M,O).
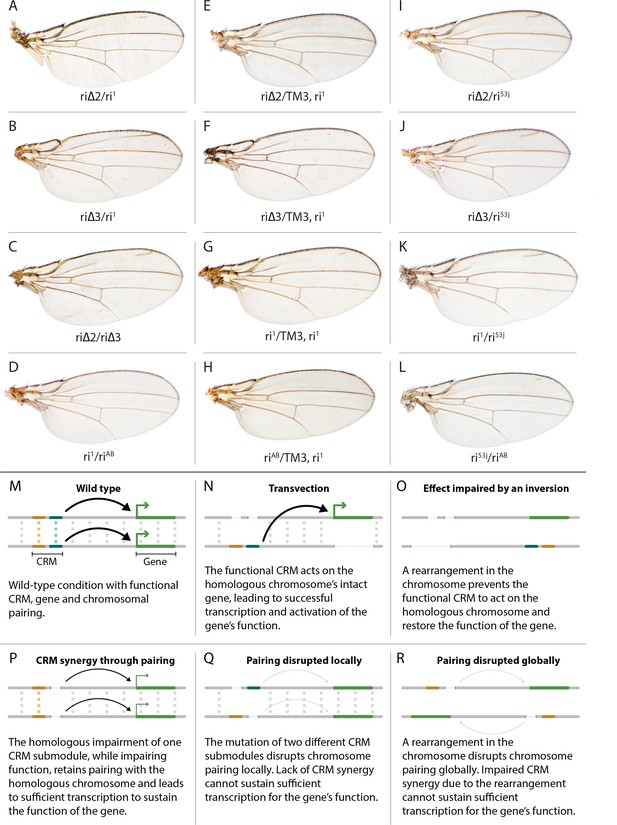
Evidence for potential chromosomal pairing dependent interactions between kni L2-CRM mutants.
(A–L) adult wings. (A) ri∆2/ri1 trans-heterozygote = kniri∆2/kniri1; (B) kniri∆3/kniri1; (C) kniri∆2/kniri∆3; (D) kniri1/kniriAB; (E) kniri∆2/TM3,kniri1; (F) kniri∆3/TM3,kniri1; (G) kniri1/TM3,kniri1; (H) kniriAB/TM3,kniri1; (I) kniri∆2/kniri53j; (J) kniri∆3/kniri53j; (K) kniri1/kniri53j; (L) kniri53j/kniriAB. (M–R) Transvection vs. CRM-synergy models: (M) Wild-type interactions between two hypothetically positively acting CRM sub-modules on homologous chromosomes. Such pairing-dependent interactions are represented generically by dotted lines between homologous CRM modules that promote the likelihood of CRM-promoter interactions (arrows). (N) Classic pairing-dependent transfection in which a functional CRM on one allele acts in-trans to promote transcription of the homologous allele. In this particular example, the functional CRM is in-cis to a loss-of-function protein coding mutation (deletion), while the functional protein coding region of the homologous chromosome is in-cis to a non-functional CRM (deletion of both sub-modules of the CRM). In this situation, all productive CRM-promoter interactions are in-trans. (O) A chromosomal rearrangement that disrupts pairing between homologous chromosomes blocks transvection. (P) When one of two redundantly acting CRM sub-modules is deleted without interfering with precise chromosomal alignment, activity can be sustained by pairing-dependent interactions between the remaining sub-modules on both homologues. (Q) If pairing between homologous CRMs is disrupted by deletion of one cooperatively acting sub-module on one allele and of the other sub-module on the homologous chromosome, the sum of the weakened activities of both non-interacting sub-modules being below the required threshold for sustaining gene function. (R) Similarly, if pairing of homologues is disrupted by a chromosomal inversion, then the two remaining isolated sub-modules do not surpass a necessary threshold for sustaining functionally sufficient transcription of the gene. For this model, we hypothesize that generic interactions between CRM modules on homologous chromosomes may modulate CRM function. Such interactions could be sustained by a variety of potential mechanisms including (1) stabilizing protein complexes acting in cis; (2) interactions between non-coding RNAs generated from one CRM and interacting with proteins, RNA, or DNA on the opposing homolog, (3) coordinated alterations in chromatin dependent on steric access to identical chromosomal sequences.
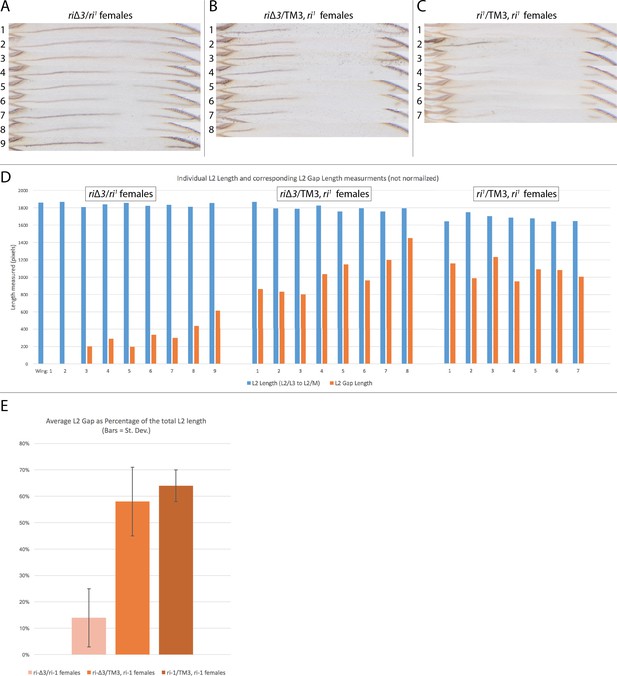
Range of riΔ3 mutant phenotypes based on chromosome pairing.
Alignment of L2 veins from a panel of flies showing the range of phenotypes associated with pairing of third chromosome homologs. (A) Phenotypes range in a configuration where pairing is presumably allowed (riΔ3/ri1); (B) Stronger vein loss phenotypes in a configuration where pairing is disrupted by multiple chromosomal inversions on the TM3 balancer chromosome (riΔ3/TM3ri1); (C) Control ri1/TM3ri1 wings showing that these alleles produce the standard ri1 mutant phenotype. (D) Graph representing the raw pixel-distance measurements of the L2 length in blue bars and gap length in orange bars. The L2 length was measured from the L2/L3 intersection to the L2/M (M = margin) intersection. (E) Average of the gap-length/L2-length ratios offering a normalized evaluation of the phenotype in the three genetic conditions. Black bars on the graph represent standard deviation.
-
Figure 3—figure supplement 1—source data 1
Raw Data for Figure 3—figure supplement 1.
Raw data of wing measurements used for evaluating the phenotype of the L2 wing vein in Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.30281.009
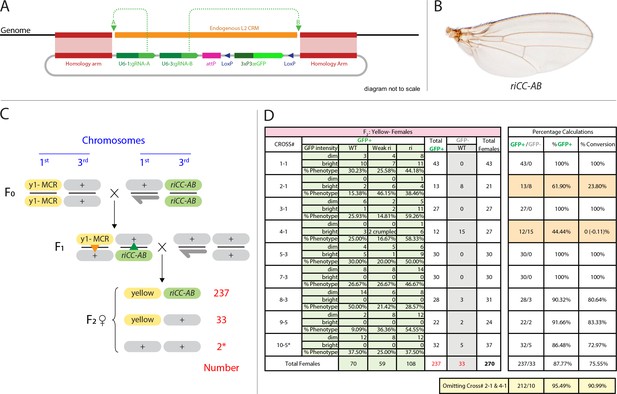
Efficient super-Mendelian transmission of a double-cut CopyCat element designed to delete the L2 CRM.
(A) Diagram of the transgenesis vector for creating the riCCAB double-cut CopyCat (CC) element, the full nomenclature for which (kniri<CC|gRNA-A,gRNA-B|3xP3-EGFP>), is denoted according the following convention that we propose for representing active genetic elements in this and future studies. We have previously outlined features of a variety of active genetic elements including: MCRs, ERACR type reversal drives, CopyCat (CC) elements, and Trans-complementing drives (TC) (Gantz and Bier, 2016). Our proposed nomenclature convention for active genetic elements uses ‘<>” to denote that the element is ‘active’: gene locus<construct type (i.e. MCR, ERACR, CC, TC)| gRNAs| dominant marker| other 5' or 3' cargo>. (B) Homozygous riCC-AB adult wing (compare to Figure 1C). (C) Crossing scheme: Homozygous y1-MCR; +/+females were crossed to homozygous +/Y; riCC-AB/riCC-AB males and resulting F1 female progeny of the genotype y1-MCR/+; riCCAB/+were then crossed to w-/Y; +/+males. F2 female progeny were then scored for inheritance of the y1-MCR element (assessed by a full body yellow- phenotype) as well as for the EGFP-marked riCC-AB element. In addition, F2 females were scored for the presence (weak or strong) or absence of an ri phenotype. (D) Table of nine crosses showing the fraction of y- females which inherited the GFP marker (number and %GFP+) and among those that did, the numbers and percentages of those with wild-type, weak ri, strong ri, or total with ri wing phenotypes, and percent of total progeny with ri phenotypes. Results from the table shown in panel (D) can be summarized as follows: In a total of nine separate crosses, transmission of the y1-MCR was observed in 270/272 female progeny (=99.3%). Two individuals derived from cross 10–5* were y+. In four of the crosses the riCC-AB element was transmitted to 100% of the progeny, in three crosses to an average of 89% of progeny, and in two crosses only Mendelian inheritance was observed (note: nearly identical percentages were observed for riCC-AB transmission in males from the same set of 9 crosses). Note that there is no correlation between the % GFP +and the % ri (weak +strong). In particular, approximately 70% of progeny exhibited some degree of ri phenotype in all four crosses that transmitted the riCCAB to 100% of their progeny as well as in the two crosses that only transmitted the riCCAB to ~50% of their progeny. These results suggest that the germline and somatic activities of this active genetic element seem separable events.
-
Figure 4—source data 1
Raw Data for Figure 4.
Raw data of phenotypical scoring of female progeny of yellow MCR / riAB-CC heterozygous female crossed to wild type males. Analyzing inheritance of copycat construct by following the GFP marker; while the MCR activity was inferred by scoring yellow body phenotypes in F2 females.
- https://doi.org/10.7554/eLife.30281.011
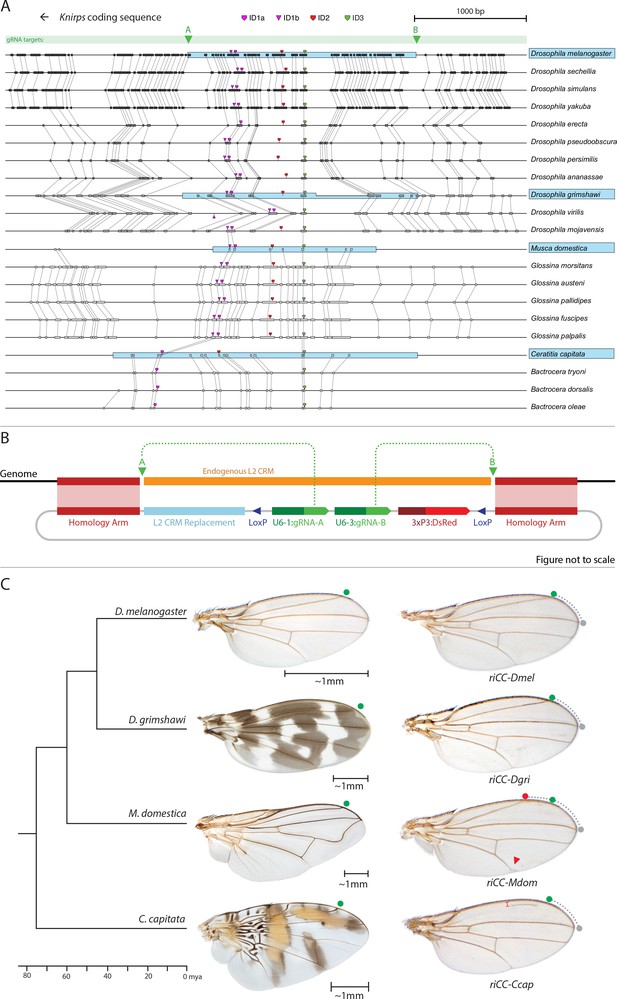
Replacement of the D. melanogaster L2-CRM with orthologous sequences from other Dipterans.
(A) Comparisons of candidate L2-CRM sequences across dipterans tested for replacement. Vertical lines connect boxes (black grey) indicating conserved identical DNA sequences. Hyper-conserved identity islands (ID sequences - shown with different colored symbols) were used to identify the L2-CMR candidate sequences (light blue boxes) in whole genome sequence BLASTS. Drosophilid species names within the different clades shown in the alignment that were tested for CRM function are boxed in blue. (B) Map of CopyCat CRM replacement vector: Diagram of plasmid used for transformation with CopyCat replacements. Elements depicted from left to right are: Homology arms (magenta boxes); replacement CRM sequences (light blue box - not drawn to scale); paired LoxP sites allowing deletion of non-replacement sequences if desired (dark blue triangle); gRNAs A and B (green arrows boxes) expressed under the control of different U6 promoters (dark green boxes); and the DsRed dominant eye color marker (red arrow) expressed under the control of the 3XP3 eye-specific promoter (maroon box) followed by 3' polyadenylation sequences derived from SV40 (not shown in diagram). Endogenous L2 CRM shown in orange (not drawn to scale with regard to CRM replacements - see panel A for proportional comparisons of CRM sizes and endpoints). (C) Replacement of the D. mel. L2-CRM with candidate orthologous sequences from other Dipterans. Approximate phylogenetic tree of Dipteran species tested on left, examples of wings from these species in the center with relative size bars and green circles indicating respective positions of the L2 vein in each species. Typical examples of vein replacement phenotypes which were highly reproducible in all cases. Note that the riCC-Dmel (full notation: kniri<CC|gRNA-A, gRNA-B|3xP3-DsRed|5'-Dmel-L2-CRM>) and riCC-Dgri (kniri<CC|gRNA-A,gRNA-B|3xP3-DsRed|5'-Dgri-L2-CRM>) replacements provide rescue of the L2 vein that is indistinguishable from the wild-type position, shown with green circles marking the wild-type position of L2 compared to L3 - dotted arc connecting to grey circles at L3 position. In riCC-Mdom (kniri< CC|gRNA-A, gRNA-B|3xP3-DsRed|5'-Mdom-L2-CRM>) wings, the L2 vein is also fully rescued but intersects the wing margin in a much anteriorly displaced position (red circle). This anterior displacement phenotype was quantitated in N = 20 wings by three separate measures: (1) ratio of the length of L2/L3 veins: average = 0.781 ± 0.006 for WT versus 0.637 ± 0.016 for riCC-Mdom, p<1.2×10−18; (2) the angle between the L2 and L3 veins diverging from their proximal intersection point: average = 13.63 ± 0.58 degrees for WT versus 18.64 ± 0.58 degrees for riCC-Mdom, p<2.0×10−17; and (3) the number of thick innervated bristles forming anterior to the point of intersection of the L2 vein with the margin: average = 11.15 ± 0.9 bristles for WT versus 28.9 ± 1.8 bristles for riCC-Mdom, p<2.8×10−20. In riCC-Ccap (kniri< CC|gRNA-A,gRNA-B|3xP3-DsRed|5'-Ccap-L2-CRM >) wings, only rescue of a partial vein segment is observed (N = 20 wings; average ratio of L2 vein segment length to that of L3 = 0.136 ± 0.03 for riCC-Ccap versus 0.781 ± 0.006 for WT, p<7.7×10−27) although its occurrence and placement are highly penetrant phenotypes. This rescued vein segment forms much closer to the margin than does the normal L2 vein in D. mel. This effect was quantitated by measuring the relative distance of the rescued vein segment from the margin at a defined PD position (arrow) relative to the distance of the L2 vein in WT at that same PD position: (N = 20 wings; average distance in riCC-Ccap rescue = 0.058 ± 0.004 mm versus 0.091 ± 0.005 mm for WT, p<6.9×10−15). Note that in both M. domestica and C. capitata wings, the L2 vein forms in a relatively more anterior position than it does in D. mel. or D. grimshawi..
-
Figure 5—source data 1
Raw data for quantifications of L2 vein phenotypes in CopyCat transformants.
Raw data of measurements and quanitifcations of relative L2 vein length to L3 vein length in 20 individual riCC-Mdom, wild type, and riCC-Ccap males; quantification of riCC-Mdom L2 vein shift by counting bristle numbers and measuring the angle of L2 to L3, and measurement of distance of L2 vein in riCC-Ccap from the margin of the wing.
- https://doi.org/10.7554/eLife.30281.013
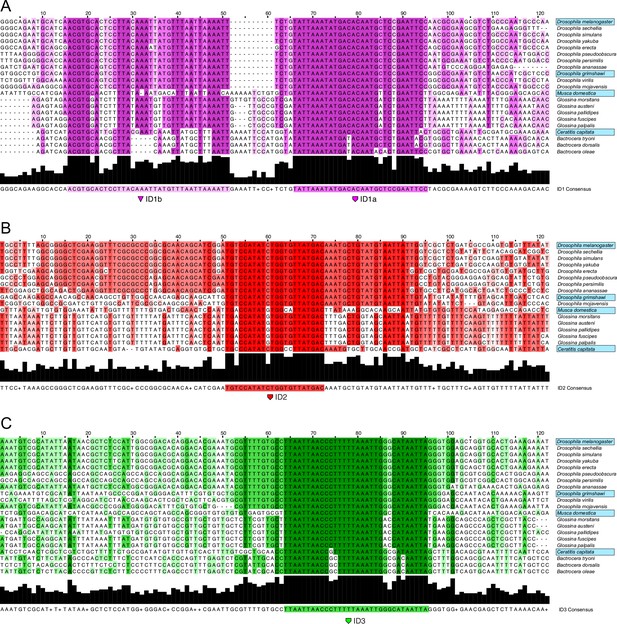
Alignment of ID sequences across Schizophora.
Nucleotide alignments of hyper-conserved identity islands across Schizophora species in clades of those species tested for activity in L2-CRM replacement experiments. Dark bar graph at the bottom of each alignment indicates percentage identity as does the differently shaded color-coding in the alignment. Species names boxed in blue were tested for CRM activity in D.mel. (A) ID1 which we split in ID1a and ID1b sub-module sequences. (B) ID2 sequences alignment. (C) ID3 sequences alignment.
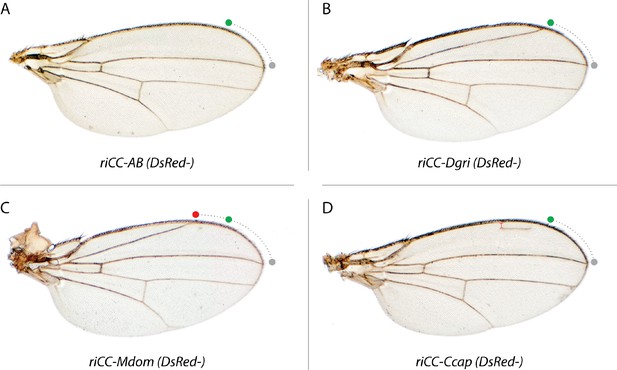
Homozygous phenotypes of CRM-only replacements.
Sequences including the Ds-Red marker gene included between the LoxP sites in the CRM replacement elements were deleted by crossing to a line expressing CRE-recombinase (see Materials and methods) and homozygous lines carrying predicted deletions of the Ds-Red cassettes were obtained. (A) D. mel. AB deletion: riCC-AB (DsRed-); (B) D. grimshawi CRM replacement: riCC-Dgri (DsRed-), (C) M. domestica CRM replacement: riCC-Mdom (DsRed-); and (D) C. capitata replacement: riCC-Ccap (DsRed-). These wings act as a control, showing that the CopyCat gRNA genes and fluorescent marker sequences had no effect to the wing phenotype when present. Note that for clarity, we label these constructs based on their parent CopyCat nomenclature but that in the absence of the DsRed and gRNA containing cassette (i.e. DsRed- derivatives) they are no longer endowed with active genetic properties.
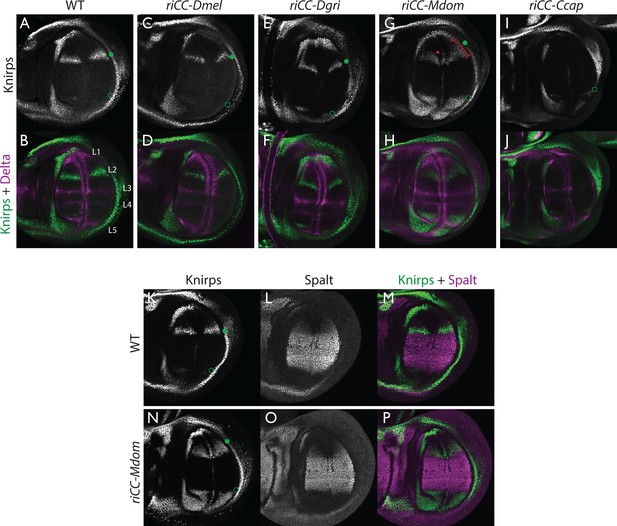
Kni expression in L2-CRM replacement lines.
Double stains of third larval instar wing imaginal discs from stocks carrying homozygous CRM replacement CopyCat elements showing expression of Kni +Delta (A–J) or Kni +Spalt (K–P). Green dot indicates approximate location of the L2 primordium and open green circles indicate posterior Sal border where a faint stripe of Kni expression is detected in wild-type D. mel wings. For panels showing double stains Kni expression is in green and Delta or Spalt expression is in magenta. Delta is a pan-vein marker expressed in the L1, L3, L4, and L5 vein primordia (labeled in panel B) and provides a positional reference for Kni expression. Spalt labels the central region of the wing where it suppresses expression of Kni in D. mel. Staining in panels showing single antibody panels alone are in black and white. (A,B) WT wing disc - see also Spalt double labels in panels (K-M; C,D) control replacement with D. mel. CRM (riCC-Dmel); (E,F) replacement with D. grimshawi candidate L2 CRM sequences (riCC-Dgri); (G,H) replacement with M. domestica candidate L2 CRM sequences (riCC-Mdom) - see also Spalt double labels in panels (N-P; I,J) replacement with C. capitata candidate L2 CRM sequences (riCC-Ccap). (K–M) Wild-type wing disc stained for Kni and Spalt. Note that Kni is expressed just anterior to cell strongly expressing Spalt. (N–P) riCC-Mdom replacement disc double stained for Kni and Spalt. Note that Kni expression is broadened relative wild-type but shares the same posterior limit as in the wild-type disc. These experiments indicate that all rescued ‘replacement’ veins form with the same posterior border bounded by strong Spalt expression as in D. mel.. Kni expression in L2 primordium of riCC-Dgri discs is normal in position and width albeit perhaps a bit weaker than with the D. mel. control replacement (riCC-Dmel). In the case of the riCC-Mdom, the rescued vein is broader than in D. mel. extending further to the anterior. In addition, there is ectopic expression along future wing margin which is absent in D. mel. or the other replacements. In the case of the riCC-Ccap element, which provides only partial rescue of the adult L2 vein, Kni expression is virtually undetectable at this stage.
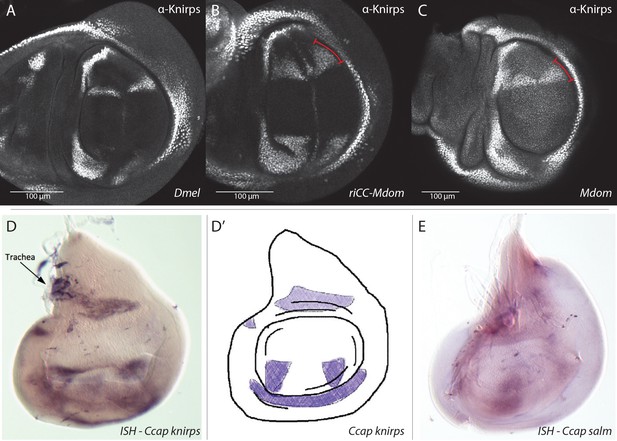
Knirps protein expression in Musca domestica and knirps (kni) mRNA and spalt-major (salm) mRNA expression in Ceratitis capitata wing imaginal discs.
(A–C) All wing discs were stained with the same anti-D.mel. Knirps antibody that was used throughout this study. Scale bars indicate relative sizes of the D.mel. versus M.dom. wing discs. (A) Wild-type D.mel. (B) D.mel. with riCC-Mdom CRM replacement. (C) Wild-type M. domestica (Mdom) wing disc. (D–E) kni and salm expression detected by in situ hybridization (ISH). (D) kni is expressed in two stripes likely corresponding to L2 (R2 +3) and maybe CuA1 or M4 (between L4 and L5?) in a C. capitata (Ccap) wing disc. (D’) diagram representing the kni expression in the C. capitata (Ccap) wing disc. (E) salm expression in a C. capitata (Ccap) wing disc. Data in (D–E) kindly provided by Natalia Siomava as part of her Ph.D. thesis under Prof. Ernst Wimmer at the University of Göttingen.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody (guinea pig) | guinea pig anti-knirps | Lunde et al. (1998) | Asian Distribution Center for segmentation antibodies: Antibody #566 | diluted 1:1000 in 1x PBS with0.1% Tween |
| Antibody (mouse) | mouse anti-engrailed | Developmental Studies Hybridoma Bank (DSHB) | RRID:AB_528194 | diluted 1:200 in 1x PBS with 0.1% Tween |
| Antibody (mouse) | mouse anti-delta | Developmental Studies Hybridoma Bank (DSHB) | RRID:AB_528224 | diluted 1:500 in 1x PBS with 0.1% Tween |
| Antibody (rabbit) | rabbit anti-spalt | de Celis et al., 1999 | diluted 1:1000 in 1x PBS with 0.1% Tween | |
| Software | Snapgene | http://www.snapgene.com/ | RRID:SCR_015052 | |
| Software | Gene palette | http://www.genepalette.org/ |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30281.018





