Reconciling isothermal titration calorimetry analyses of interactions between complexin and truncated SNARE complexes
Figures
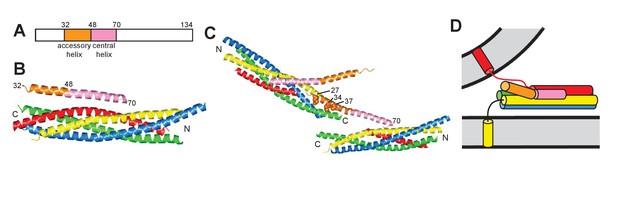
Models of the inhibitory function of Complexin.
(A) Domain diagram of CpxI. Selected residue numbers are indicated above the diagram. (B) Ribbon diagram of the crystal structure of the SNARE complex bound to CpxI(26-83) (PDB code 1KIL) (Chen et al., 2002). Synaptobrevin is colored in red, syntaxin-1 in yellow, SNAP-25 in blue and green (N-terminal and C-terminal SNARE motifs, respectively), and CpxI(26-83) in orange (accessory helix) and pink (central helix). N and C indicate the N- and C-termini of the SNARE motifs. Selected residue numbers of CpxI(26-83) are indicated. (C) Ribbon diagram of the crystal structure of the SNAREΔ60 complex bound to the CpxI(26-83) superclamp mutant (PDB code 3RK3) (Kümmel et al., 2011). Two complexes are shown to illustrate the zigzag array present in the crystals. Selected residue numbers are indicated for one of the scCpxI(26-83) molecules, which binds to one SNAREΔ60 complex through the central helix and to another SNAREΔ60 complex through the accessory helix. The three mutated residues in the accessory helix are shown as spheres and their residue numbers are indicated. (D) Model postulating that the Complexin accessory helix inhibits neurotransmitter release because of steric repulsion with the vesicle membrane. The model is based on the crystal structure shown in (A), but assumes that the C-terminus of the synaptobrevin SNARE motif is not assembled into the SNARE complex. This figure is based on Figure 1 of Trimbuch et al. (2014), with modifications.
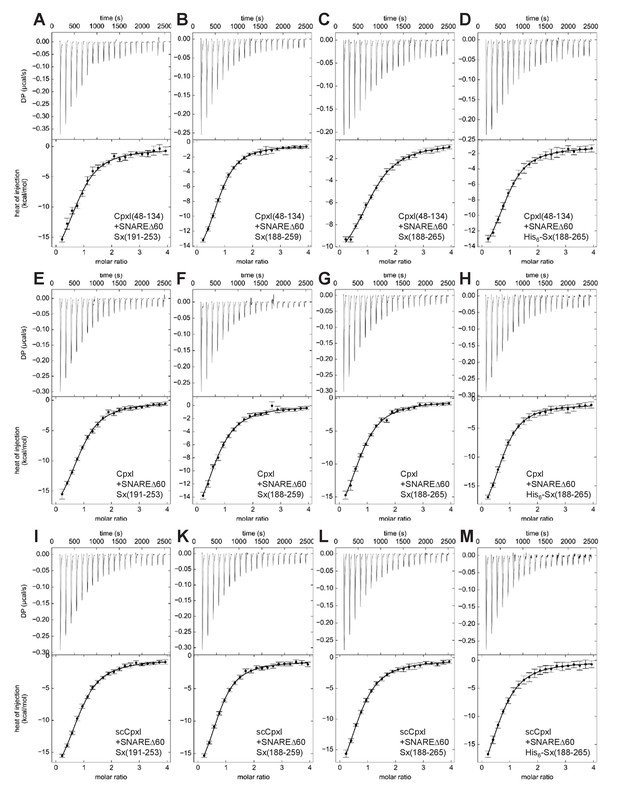
ITC analysis of CpxI-SNAREΔ60 interactions by direct titration.
The various panels show direct titrations of SNAREΔ60 containing syntaxin-1(191–253) (A,E,I), syntaxin-1(188–259) (B,F,K), syntaxin-1(188–265) (C,G,L) or His6-syntaxin-1(188–265) (D,H,M) with CpxI(48-134) (A–D), CpxI (E–H) or scCpxI (I–M), monitored by ITC. The upper panels show the baseline- and singular-value-decomposition-corrected thermograms for the respective experiments. The circles in the lower panels are the integrated heats of injection, with the error bars representing estimated errors for these values (Keller et al., 2012). The lines in these panels represent the respective fits of the data to a single binding site ‘A + B <->AB’ model.
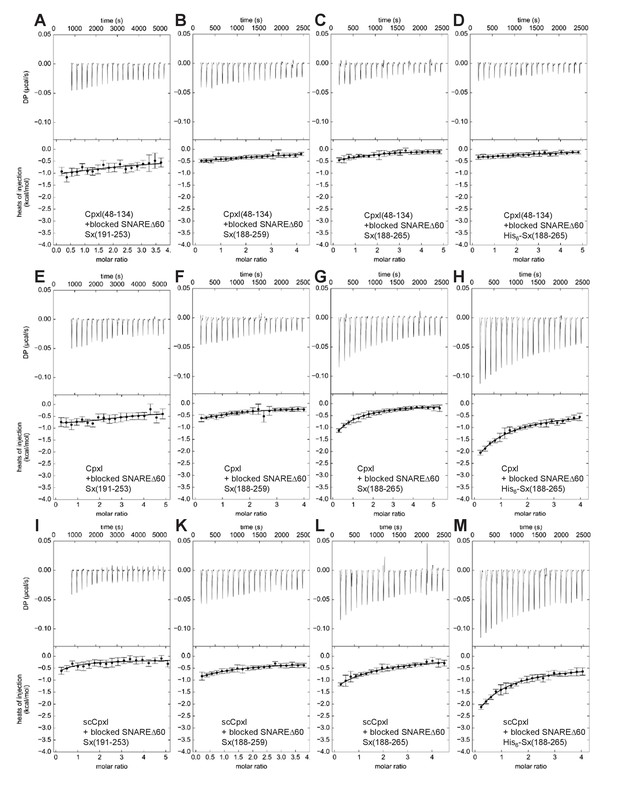
ITC analysis of CpxI-SNAREΔ60 interactions through blocking assays.
The various panels show blocking assays monitored by ITC where SNAREΔ60 complex blocked with 4.9 equivalents of CpxI(48-134) and containing syntaxin-1(191–253) (A,E,I), syntaxin-1(188–259) (B,F,K), syntaxin-1(188–265) (C,G,L) or His6-syntaxin-1(188–265) (D,H,M) was titrated with CpxI(48-134) itself (A–D), CpxI (E–H) or scCpxI (I–M). The upper panels show the baseline- and singular-value-decomposition-corrected thermograms for the respective experiments. The circles in the lower panels are the integrated heats of injection, with the error bars representing estimated errors for these values (Keller et al., 2012). The lines in these panels represent the respective fits of the data to a single binding site ‘A + B < ->AB’ model, but note that no meaningful thermodynamic parameters can be derived from these data sets.
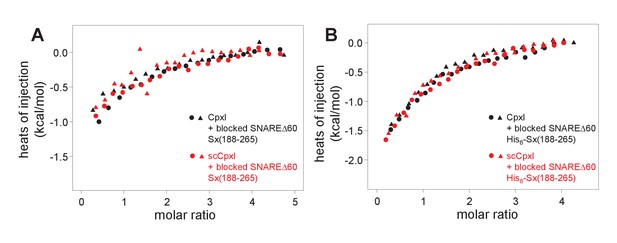
The superclamp mutation does not alter the heat release observed in the blocking assays.
The plots show superpositions of ITC data obtained in blocking assays such as those described in Figure 3, including two separate experiments performed with WT CpxI and two separate experiments performed with scCpxI, titrated into blocked SNAREΔ60-Sx265 (A) or His6-SNAREΔ60-Sx265 (B) complexes. To facilitate comparison of the four data sets shown in each panel, a constant value resulting from the average of the last five data points was subtracted from each data point of a given dataset. Note that, as a consequence, the baseline at the end of the titration is closer to zero than observed in the plots of Figure 3G,H,L,M, where the baseline was determined by the data fitting procedure. The data superpositions show that the superclamp mutation does not markedly influence the heat release observed in experiments performed with blocked SNAREΔ60-Sx265 (A) or His6-SNAREΔ60-Sx265 (B) complexes.
Tables
Summary of KDs (in μM units) between CpxI proteins and SNAREΔ60 complexes containing different syntaxin-1 fragments measured by ITC*.
https://doi.org/10.7554/eLife.30286.006| SNAREΔ60-Sx253 | SNAREΔ60-Sx259 | SNAREΔ60-Sx265 | His6-SNAREΔ60-Sx265 | |
|---|---|---|---|---|
| CpxI(48-134) | 2.0 [1.4–2.8] | 1.4 [1.2–1.7] | 2.0 [1.6–2.5] | 1.8 [1.5–2.1] |
| CpxI | 1.9 [1.6–2.3] | 2.5 [2.0–3.1] | 2.4 [2.0–3.0] | 2.2 [1.8–2.6] |
| scCpxI | 2.0 [1.8–2.4] | 2.2 [1.7–2.5] | 2.2 [1.9–2.6] | 2.2 [1.9–2.5] |
-
*At least two independent experiments were performed for each combination of CpxI protein and SNAREΔ60 complex. KDs were derived from global fit of the independent experiments performed for each combination. For all KDs, 68.3% confidence intervals calculated using the error-surface projection method are indicated between brackets.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30286.007





