Mechanical force induces mitochondrial fission
Figures
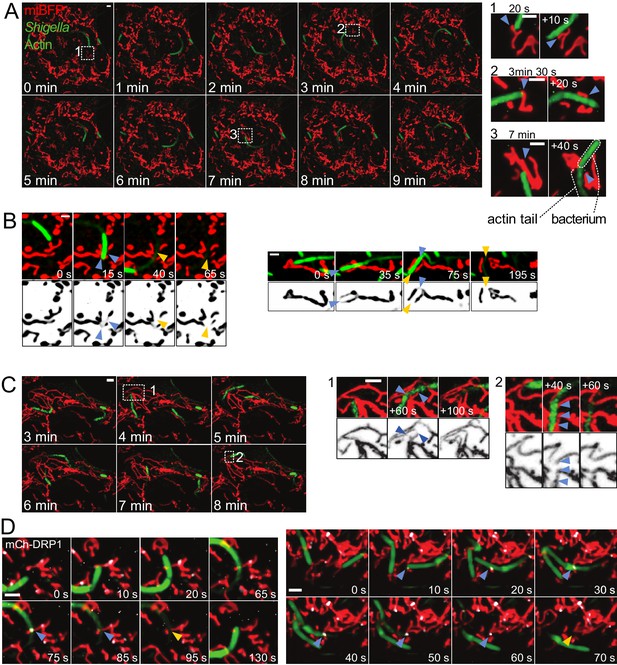
Mitochondria undergo DRP1-mediated fission upon encountering actin-propelled Shigella.
(A) U2OS KERMIT cells (stably expressing mtBFP) were transfected with mCherry-Lifeact plasmid and infected with mCherry-labelled S. flexneri. Left, overview of time-lapse microscopy results presented at 1 min interval. Right, magnifications of selected areas from Left at the indicated times. Note that the times in the right inset do not necessarily match the times in the overview on the left. Arrowheads indicate mitochondria positions before and after impact with bacterium. (B) COS7 cells transduced with lentiviruses expressing GFP-Lifeact and mtBFP were infected with GFP-labelled S. flexneri. Imaging was performed as in A. Shown are four individual fission events upon encounter with S. flexneri. Blue and orange arrowheads indicate mitochondria before and after fission, respectively. (C) DRP1CRISPR U2OS KERMIT cells were subjected to the same treatment and analysis as in A. Numbered boxes as in A. Blue arrowheads, thinning mitochondrial tubules due to impact by S. flexneri, followed by recovery of mitochondrial tubules without fission. (D) mCherry-DRP1-expressing U2OS cells were treated as in B. Blue arrowheads, recruitment of DRP1 (white) at sites of encounter with S. flexneri. Orange arrowheads, subsequent fission events. Scale bars, 2 µm.
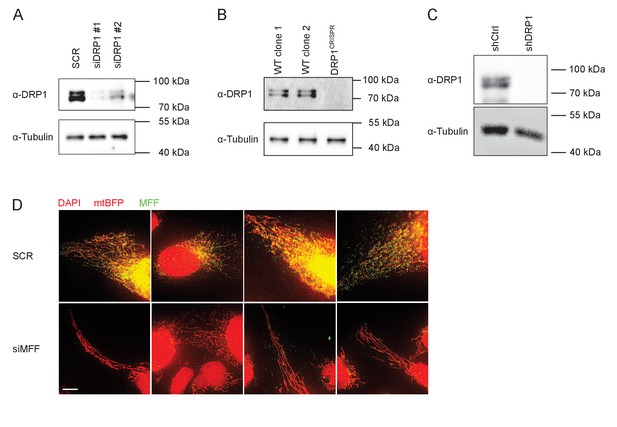
All detectable forms of DRP1 are depleted by siRNA, shRNA or CRISPR-induced mutations.
(A) KERMIT cells were transfected three times consecutively within 72 hr, each with 40 nM of scrambled (SCR) or DRP1 siRNA #1 or #2. 40 µg of total lysate was loaded per lane onto a 10% polyacrylamide gel and subjected to SDS-PAGE and immunoblotting. Tubulin immunoblot is shown as loading control. (B) Total lysates were prepared from KERMIT cells (WT clone 1), from a second clone derived from U2OS cells (WT clone 2) and from DRP1CRISPR KERMIT cells. 20 µg of lysate per sample was analyzed on 10% polyacrylamide gel and subjected to SDS-PAGE and immunoblotting. Proteins were detected using specific antibodies. Tubulin immunoblot is shown as loading control. (C) U2OS mtBFP cells were incubated with either virus encoding shRNA targeted against DRP1 or a virus transduction control overnight at 37°C. Subsequently the medium was exchanged and puromycin (1 µg/ml) was added. Cells were grown under selection for four days and subsequently prepared for SDS-PAGE and immunoblotting. Equal amounts of protein were loaded and proteins were detected using specific antibodies. Tubulin immunoblot is shown as loading control. (D) KERMIT cells were transfected with 10 nM of scrambled (SCR) or MFF-targeting siRNA. Cells were fixed 2 days post transfection using 4% paraformaldehyde and stained with DAPI and MFF-specific antibody. All images were acquired using the same excitation and acquisition settings, and presented at the same intensity settings. Scale Bar, 10 µm.
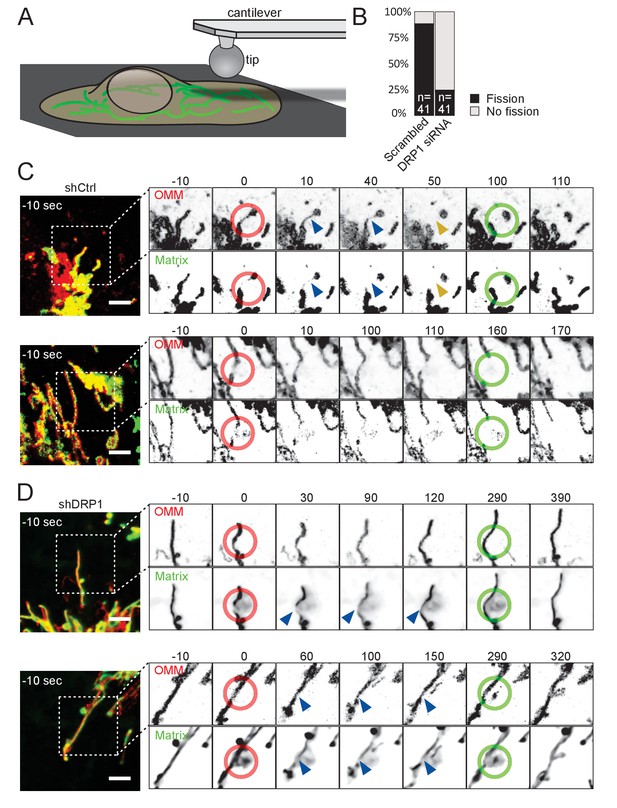
Mitochondria undergo DRP1-dependent fission upon AFM-mediated force application.
(A) Scheme of the experimental setup. (B) Quantification of fission events elicited by the application of AFM-induced mechanical forces observed in cells treated either with scrambled or DRP1 siRNA. Successful force application to an individual mitochondrion was defined as visible constriction of the mitochondrial matrix following tip approach (examples shown with blue arrowhead in C and D). (C) U2OS cells stained with Mitotracker Deep Red and transduced with viruses encoding mCherry-FIS1TM and a control shRNA were imaged by time-lapse microscopy. Two examples are shown. At t = 0 s, the cantilever of the AFM approached the cell in Contact mode, with a force set at 15 nN, at the position of the red ring. Green rings mark the time and area of tip retraction. Blue arrowheads indicate mitochondria that are visibly thinned by the pressure but have not yet undergone fission. Fission events are indicated by an orange arrowhead. OMM panels, mCherry-FIS1TM (red). Matrix panel, mtBFP (green). Scale bar, 5 µm. (D) As in (C) except that the cells were treated with a virus encoding DRP1 shRNA. Scale bar, 5 µm.
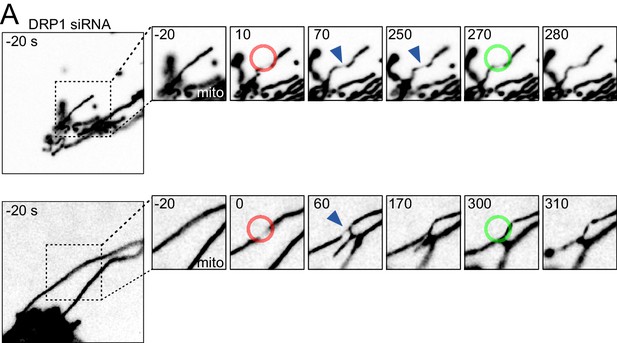
Mitochondria undergo DRP1-dependent fission upon AFM-mediated force application.
(A) U2OS KERMIT cells stably expressing mtBFP and Sec61β-GFP and treated with DRP1 siRNA were imaged by time-lapse microscopy. Two examples are shown. At t = 0 s, the cantilever of the AFM approached the cell in Contact mode, with a force set at 15 nN, at the position of the red ring. Green rings mark the time and area of tip retraction. Blue arrowheads indicate mitochondria that are visibly thinned by the pressure. Mito panels, mtBFP. Scale bar, 5 µm.
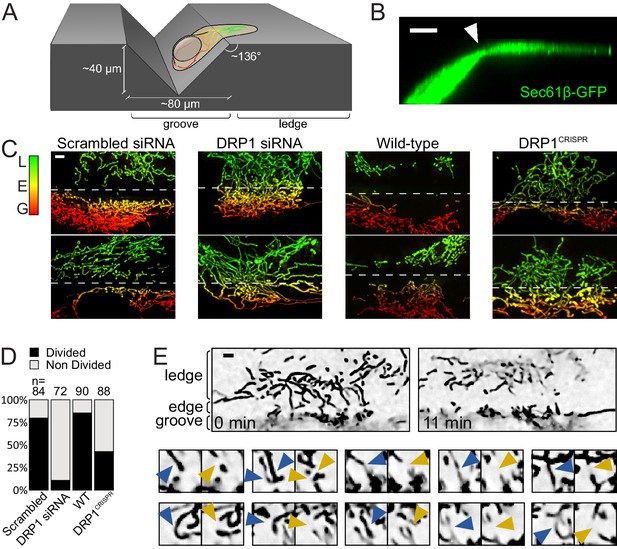
Mitochondrial fission upon cell deformation.
(A) Scheme of the experimental setup. (B) 3D projection of a U2OS cell expressing Sec61β-GFP showing thinning of the cytoplasm at the groove’s edge (arrowhead). (C) Mitochondria of indicated cells grown on vinyl records. Mitochondria are color-coded according to their Z-position (red in the groove, green on the ledge). The dashed lines indicate the approximate position of the edge. (D) Quantification of the number of cells showing a divided mitochondrial network, defined as having no mitochondria spanning the edge between the groove and the ledge. Number of cells analyzed are indicated on each bar. (E) Time-lapse microscopy of the fission events leading to divided mitochondrial network. Top panel, low magnification of the start- and end-points of the recording. Lower panels, individual fission events captured during the time course. Blue arrowheads, mitochondria before fission; Orange arrowheads, mitochondria after fission. Scale bars, 5 µm.
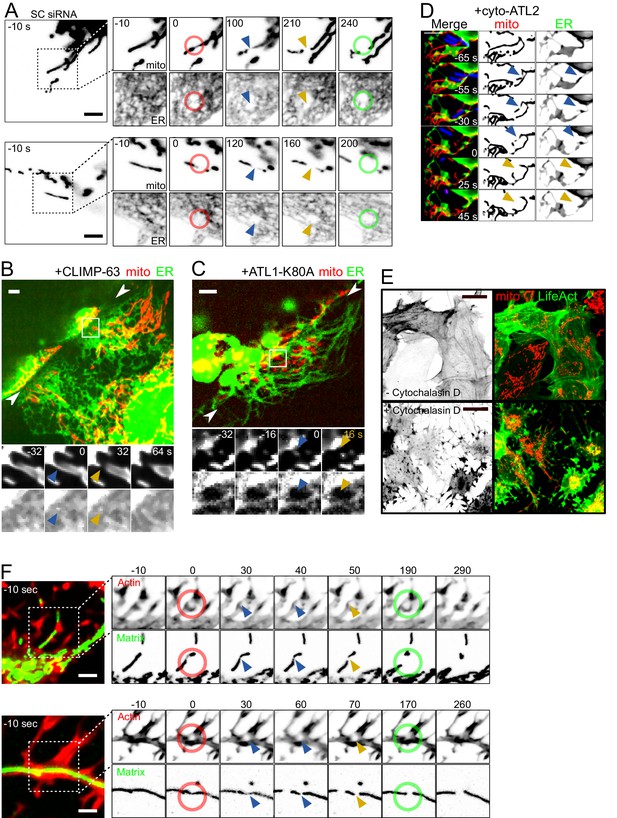
Force-induced mitochondrial fission upon ER dynamics perturbations.
(A) U2OS KERMIT cells stably expressing mtBFP and Sec61β-GFP and treated with scrambled siRNA were imaged by time-lapse microscopy. Two examples are shown. At t = 0 s, the cantilever of the AFM approached the cell in Contact mode, with a force set at 15 nN, at the position of the red ring. Green rings mark the time and area of tip retraction. Blue arrowheads indicate mitochondria that are visibly thinned by the pressure but have not yet undergone fission. Fission events are indicated by an orange arrowhead. (B) Cells were seeded on vinyl records and transfected with a CLIMP-63-overexpressing plasmid, effectively converting large fractions of the ER to sheets. Mitochondria underwent fission when spanning over the edge of the groove whether or not the ER had been converted to sheets at the site of fission. The two facing arrowheads indicate the position of the groove’s edge. (C) As in (B) but cells were instead transfected with a construct expressing a dominant-negative form of ATL1-K80A, which inhibits ER interconnection and increases the size of gaps in the ER network. As a result, mitochondria can be observed undergoing fission at sites devoid of ER while spanning the edge of the groove. (D) U2OS KERMIT cells were transduced with a lentivirus expressing RFP-Lifeact (blue) and transfected with a cyto-ATL2 expression plasmid. Cells were then infected with mCherry-labelled S. flexneri (blue). Mitochondria can be observed undergoing fission at sites stimulated by motile bacterium. The blue and yellow arrowheads represent mechanically constricted sites before and after fission, respectively. Mito panels, mtBFP. ER panel, Sec61β-GFP. Scale bars 5 µm. (E) U2OS cells transduced with GFP-Lifeact and matrix-targeted RFP were treated with 5 µg/µl of Cytochalasin D for 90 min. (F) U2OS cells transduced with GFP-Lifeact and matrix-targeted RFP were treated for 90 min with 1 µg/µl (upper panel) or 5 µg/µl Cytochalasin D (lower panel), respectively. At t = 0 s, the cantilever of the AFM approached the cell in Contact mode, with a force set at 15 nN, at the position of the red ring. Green rings mark the time and area of tip retraction. Blue arrowheads indicate mitochondria that are visibly thinned by the pressure but have not yet undergone fission. Fission events are indicated by an orange arrowhead.

Control for INF2 knockdown efficiency.
U2OS cells stably expressing mitochondrial matrix-targeted BFP (mtBFP) were reverse-transfected with scrambled (SCR) siRNA or siRNA specifically targeting INF2-CAAX isoform. A day later all cells were transfected with a plasmid expressing GFP-INF2-CAAX. 48 hr post siRNA transfection cells were fixed and imaged on a DeltaVision epifluorescence microscope. Scale bar, 5 µm.
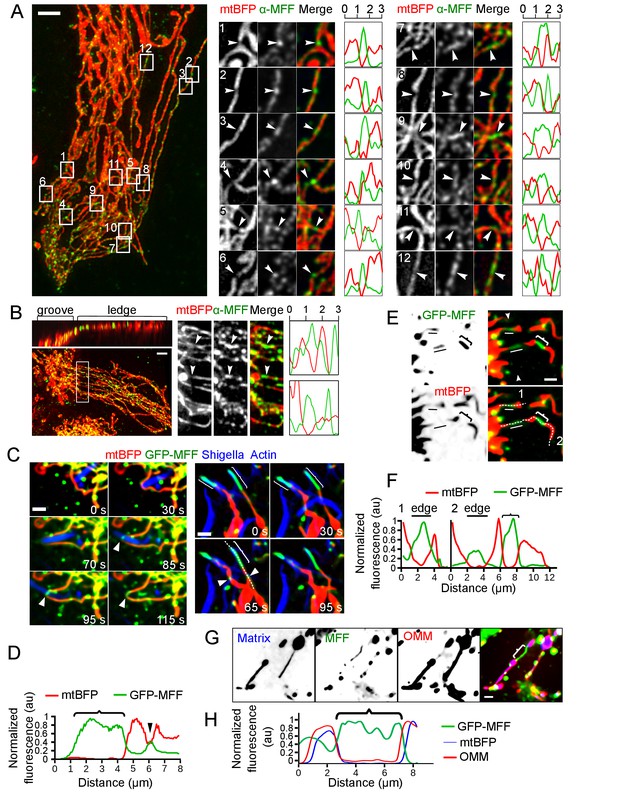
MFF recruitment to mechanically strained sites.
(A) Immunofluorescence of Kermit cells transduced with shDRP1, using an anti-MFF antibody (green). Mitochondrial matrix (mtBFP) is shown in red. Insets on the right correspond to the framed areas on the left. Arrowheads point at naturally occuring constrictions on the mitochondria. Plots are linescans of the mitochondria (red) and MFF (green) signals around the constriction. X-axis is in µm. Y-axis is normalized fluorescence in arbitrary units. (B) as in A, but the cells were grown on gramophone records. Top left panel is a 3D projection of the same cell rotated by 90° around the x-axis. The white box on the left corresponds to the magnified area on the right. Arrowheads indicate mitochondrial constrictions at the groove’s edge. (C) U2OS cells were transduced with lentiviral vectors expressing mtBFP, RFP-Lifeact, GFP-MFF as well as shRNA targeting DRP1. Puromycin-resistant cells (shRNA-positive) were then infected with RFP-labeled S. flexneri, and imaged by time-lapse microscopy. MFF is recruited to sites of encounter with S. flexneri (white arrowheads). Right panels also show two examples of MFF enrichment at sites of mitochondria thinning (curly brackets), as indicated by reduction of matrix mtBFP signal, independent of Shigella encounter. (D) Line scan of mtBFP and MFF signal of the white dotted line in (C). Arrowhead and curly bracket correspond to same zones in (C). Normalized background-subtracted pixel values are plotted as arbitrary units. (E) Z-projected image of a transduced U2OS cell spanning the edge of vinyl groove expressing mtBFP, GFP-MFF as well as shRNA targeting DRP1. The groove’s edge is indicated by two facing arrowheads on top right panel. Two stabilized individual mitochondrial tubules span over the edge (white line), and show loss of matrix BFP signal and increased MFF signal. Another example of GFP-MFF enrichment to a constricted mitochondrial tubule outside of the edge area is indicated by a curly bracket. (F) Dotted white lines 1 and 2 in (E) are selected for line plots as in (D). (G) U2OS cells were transduced with lentiviral vectors expressing mtBFP, mCherry-Fis1TM (OMM), as well as shRNA targeting DRP1. Puromycin-resistant cells (shRNA-positive) were then transfected with GFP-MFF, and imaged by time-lapse microscopy. MFF spontaneously stabilizes thin mitochondrial section (curly brackets) that are devoid of matrix staining but retain continuous OMM signal. (H) Line scan of mtBFP, GFP-MFF and OMM signal of the curly bracket in (G). Scale bars, A-B, 5 µm, C-H 2 µm.
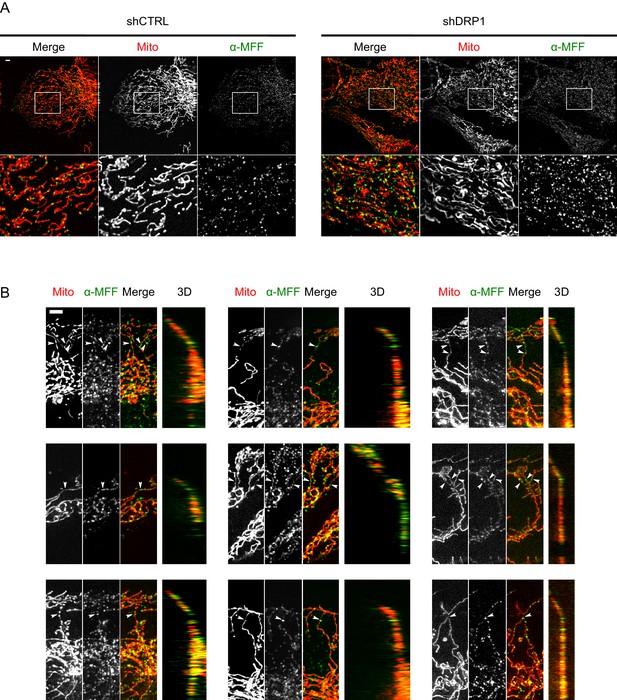
MFF immunostaining in WT and DRP1-deficient cells.
(A) KERMIT cells were transduced with lentiviruses expressing either a non-targeting shRNA (shCTRL) or a DRP1-targeting shRNA. Cells were then cultured under puromycin selective pressure for 10 days before they were fixed, immunostained using a MFF-specific antibody, and imaged on a DeltaVision epifluorescence microscope. Scale bar, 5 µm. (B) More examples of cells grown of gramophone records and imaged as in Figure 5B. Arrowheads indicate mitochondrial constrictions at grooves’ edges. For each example, the rightmost panel is a 3D projection of the image rotated by 90° about the Y-axis.
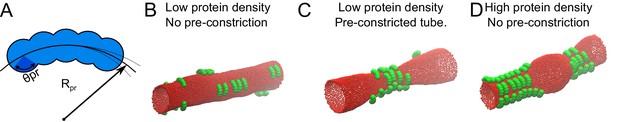
Monte Carlo simulation of protein-membrane interactions for different conditions.
(A) Proteins were modelled as a linear chain built out of five spheres positioned on a circular arc. Each sphere has a radius of σ. The center-to-center distance between the spheres within a protein was adjusted to 2/3 of σ. Rpr, is the radius of the arc, for which we found the optimal value to be 3.5σ (see supplement), and θpr is the protein-membrane contact angle, which we set to π/4, in order for the proteins to attract the membrane only by the inner part of their structure. (B) 20 proteins as in A with an optimized Rpr were allowed to reach equilibrium on a membrane tube with a radius of 10σ and a length of 100σ. (C) 20 proteins as in A were simulated on a membrane tube with a pre-constriction (radius at the center of the constriction = 3σ) and allowed to equilibrate. (D) 50 proteins as in A were allowed to reach equilibrium on a membrane tube as in B (without pre-constriction).
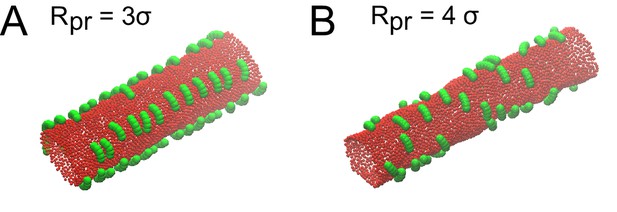
Optimization of the binding curvature of the protein during Monte Carlo simulation.
Proteins were modelled as in Figure 6D. Proteins with different radii of curvatures Rpr were allowed to reach equilibrium on a membrane tube with a radius of 10σ and a length of 100σ. Proteins adopted different assembly behaviors at different radiuses. (A) At Rpr = 3 σ, proteins attracted each other, but only in the longitudinal, and not angular, direction. (B) At Rpr = 4 σ, proteins did not self-assemble at all.
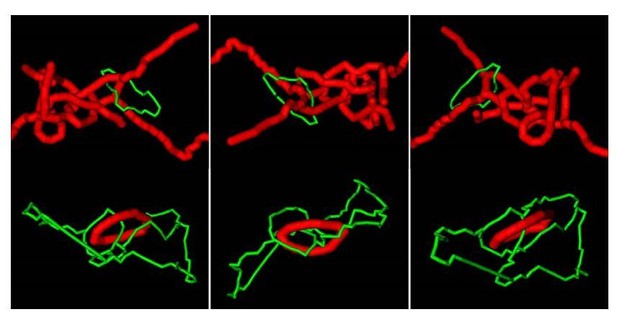
Concatenations between the ER and mitochondria in the absence of mitochondrial fission machinery.
Two examples of concatenations observed in DRP1 shRNA-treated U2OS KERMIT cells. The relevant parts of the ER (green) and mitochondria (red) were traced manually and 3D reconstituted with Fiji. These representations do not faithfully represent the morphology of the organelles, but show their relative topology, highlighting concatenations.
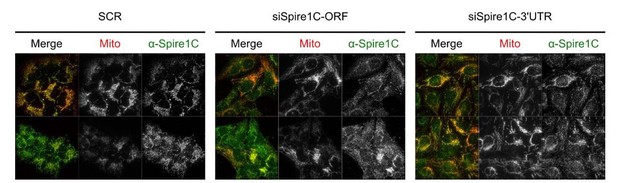
Immunostaining of Spire1C in cells treated with scrambled or Spire1C-targeting siRNAs.
U2OS cells expressing mitochondrial matrix-targeting BFP (Mito) were transfected with either scrambled (SCR) or two siRNAs targeting either the ORF or 3’ UTR of Spire1C. Cells were fixed 72 hours post transfection and immunofluorescence staining was performed using the Exon C antibody as previously described (Manor et al., 2015). Samples were then imaged using a 60x oil objective (1.4NA DIC Oil PlanApoN) on a DeltaVision microscope. All images were taken using the same settings and presented at the same intensity levels.
Videos
Mitochondria are pushed aside upon impact with Shigella.
U2OS KERMIT cells (stably expressing mtBFP) were transfected with mCherry-Lifeact plasmid and infected with mCherry-labelled S. flexneri. Red, mitochondria. Green, Shigella and actin. Arrowheads indicate events where mitochondrial tubules make way for Shigella upon encounter. Scale bar, 2 µm. This movie relates to Figure 1A.
Mitochondria divide upon encounter with Shigella.
Cos7 cells transduced with lentiviruses expressing mtBFP and GFP-Lifeact were infected with GFP-labelled S. flexneri. Red, mitochondria. Green, Shigella and actin. Blue and orange arrowheads indicate mitochondria before and after Shigella -induced fission, respectively. Scale bar, 2 µm. This movie relates to Figure 1B.
Disturbance to mitochondrial morphology by Shigella in DRP1CRISPR knockout cells.
DRP1CRISPR knockout U2OS KERMIT cells (stably expressing mtBFP) were transfected with mCherry-Lifeact plasmid and infected with mCherry-labelled S. flexneri. Red, mitochondria. Green, Shigella and actin. Arrowheads indicate thinning mitochondrial tubules due to impact by Shigella. One blue arrowhead (at 860 s) points to a DRP1-independent fission event that occurs away from the Shigella impact point. Scale bar, 2 µm. This movie relates to Figure 1C.
DRP1 recruitment and subsequent mitochondrial fission upon encounter with Shigella.
U2OS cells transduced with lentiviruses expressing GFP-Lifeact and mtBFP were transfected with mCherry-DRP1 plasmid and infected with GFP-labelled S. flexneri. Red, mitochondria. Green, Shigella and actin. White, DRP1. Blue and orange arrowheads indicate mitochondria before and after Shigella-induced fission, respectively. Scale bar, 2 µm. This movie relates to Figure 1D.
DRP1 recruitment and subsequent mitochondrial fission upon encounter with Shigella.
As in Video 4. Blue arrowheads indicate event where Shigella crosses a mitochondria region that was already coated with low level of DRP1. Orange arrowheads indicate formation of a bright DRP1 focus at this site, which subsequently undergoes fission. Scale bar, 2 µm.
Mitochondrial fission induced by contact with an AFM tip.
U2OS cells stained with Mitotracker Deep Red and transduced with viruses encoding mCherry-FIS1TM and a control shRNA were imaged by time-lapse microscopy. Six examples are shown. The right panel is a magnification of the box in the left panel. At t = 0, force was applied approximately at the center of the red ring. Force was released by AFM tip retraction at time points when the red ring turns green. Blue and yellow arrowheads mark the constricted mitochondria before and after fission respectively. This movie relates to Figure 2C. Scale bar, 2 µm.
Mitochondria in DRP1-deficient cells mechanically stressed by contact with an AFM tip.
As in Video 6 except that cells were treated with virus encoding DRP1 shRNA. This movie relates to Figure 2D. Scale bar, 2 µm.
Mitochondria in DRP1-deficient cells mechanically stressed by contact with an AFM tip.
This movie relates to Figure 2—figure supplement 1.
90-degree tilting of a 3D-reconstructed KERMIT cell expressing Sec61β-GFP, grown on a vinyl record.
This movie relates to Figure 3B.
Time-lapse recording of a KERMIT cell cultured on a vinyl record.
The groove and ledge areas are indicated at the beginning and the end of the movie. In the second part of the movie, fission events are indicated with arrowheads (blue, before fission; orange, after fission). This movie relates to Figure 3E.
The AFM tip displaces the ER during force-induced mitochondral fission.
U2OS KERMIT cells stably expressing mtBFP and Sec61β-GFP and treated with scrambled siRNA were imaged by time-lapse microscopy. Three examples are shown. At t = 0 s, the cantilever of the AFM approached the cell in Contact mode, with a force set at 15 nN, at the position of the red ring. Green rings mark the time and area of tip retraction. Blue arrowheads indicate mitochondria that are visibly thinned by the pressure but have not yet undergone fission. Fission events are indicated by an yellow arrowhead. This movie relates to Figure 4A.
Force-induced mitochondral fission in cells overexpressing CLIMP-63.
KERMIT cells were seeded on vinyl records and transfected with a CLIMP-63-overexpressing plasmid. Fission events along the groove edge are indicated with arrowheads (blue, before fission; orange, after fission). Blue dashed line at the beginning of each video indicates the position of the edge. Time stamp is relative to the first fission event in each movie. This video relates to Figure 4B.
Force-induced mitochondral fission in cells overexpressing ATL1-K80A.
KERMIT cells were seeded on vinyl records and transfected with an ATL1-K80A-overexpressing plasmid. Fission events along the groove edge are indicated with arrowheads (blue, before fission; orange, after fission). Blue dashed line at the beginning of each video indicates the position of the edge. Time stamp is relative to the first fission event in each movie. This video relates to Figure 4C.
Shigella-induced mitochondral fission in cells overexpressing cyto-ATL2.
U2OS KERMIT cells were transduced with a lentivirus expressing RFP-Lifeact (blue) and transfected with a cyto-ATL2 expression plasmid. Cells were then infected with mCherry-labelled S. flexneri (blue). Mitochondria can be observed undergoing fission at sites stimulated by motile bacterium. Blue and yellow arrowheads represent mechanically constricted sites before and after fission, respectively. Mito panels, mtBFP. ER panel, Sec61β-GFP. Scale bars, 2 µm. This video relates to Figure 4D.
INF2-independent mitochondrial fission induced by Shigella.
U2OS cells transduced with lentiviruses expressing mtBFP and GFP-Lifeact were reverse-transfected with scrambled (SCR) siRNA or siRNA specifically targeting INF2-CAAX isoform. 72 hr later, they were infected with GFP-labelled S. flexneri. Red, mitochondrial matrix. Green, Shigella and actin. Blue and orange arrowheads indicate mitochondria before and after Shigella -induced fission, respectively. Scale bar, 2 µm.
Actin-independent force-induced mitochondral fission.
U2OS cells transduced with GFP-Lifeact and matrix-targeted RFP were treated for 90 min with Cytochalasin D (Examples 1, 3, 5 at 1 µg/µl; Examples 2, 4 at 5 µg/µl). At t = 0 s, the cantilever of the AFM approached the cell in Contact mode, with a force set at 15 nN, at the position of the red ring. Green rings mark the time and area of tip retraction. Blue arrowheads indicate mitochondria that are visibly thinned by the pressure but have not yet undergone fission. Fission events are indicated by an orange arrowhead. This video relates to Figure 4F.
MFF recruitment to mitochondria constricted by the encounter with Shigella.
U2OS cells transduced with lentiviruses expressing RFP-Lifeact, mtBFP, GFP-MFF and DRP1-specific shRNA were infected with RFP-labelled S. flexneri. Red, mitochondria. Blue, Shigella and actin. Green, GFP-MFF. Arrowheads indicate MFF recruitment to constricted mitochondria. This movie relates to Figure 5C.
High level MFF overexpression stabilizes thin, matrix-free mitochondria.
U2OS cells were transduced with lentiviral vectors expressing mtBFP, mCherry-Fis1TM (OMM), as well as shRNA targeting DRP1. Puromycin-resistant cells (shRNA-positive) were then transfected with GFP-MFF, and imaged by time-lapse microscopy. MFF spontaneously stabilizes thin mitochondrial section (white arrowheads) that are devoid of matrix staining but retain continuous OMM signal. This movie relates to Figure 5G.
Monte Carlo simulation of low density of protein on a membrane tube.
Proteins and membranes were modelled as in Figure 6. 20 proteins with an Rpr of 3.5 σ were allowed to reach equilibrium on a membrane tube with a radius of 10 σ and a length of 100 σ.
Monte Carlo simulation of low density of protein on a membrane tube with a pre-constriction site.
Proteins and membranes were modelled as in Figure 6 and simulated as in Video 19, except that a pre-constricted site was present on the membrane tube throughout the simulation.
Monte Carlo simulation of high density of protein on a membrane tube.
Proteins and membranes were modelled as in Figure 6 and simulated as in Video 19, except that 50 instead of 20 proteins were present on the membrane tube.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (Shigella flexneri,serotype 5a strain M90T) | GFP-tagged shigella | other | Jost Enninga (Paris) | |
| strain, strain background (Shigella flexneri,serotype 5a strain M90T) | RFP-tagged shigella | other | Jost Enninga (Paris) | |
| strain, strain background (Shigella flexneri,serotype 5a strain M90T) | mCherry-tagged shigella | PMID:24039575 | strain only used in S. Mostowy lab (London) | |
| cell line (Homo sapiens) | U2OS | other | Matthias Peter (Zurich) | |
| cell line (H. sapiens) | U2OS-KERMIT | PMID:26259702 | ||
| cell line (Cercopithecus aethiops) | COS7 | UCSF cell culture facility | ||
| cell line (H. sapiens) | KERMIT-DRP1CRISPR | this paper | CRISPR-mediated DRP1 knockout cell line, generated using pX330-DRP1e × 2 and pX330- DRP1e × 6 (see entry for these plasmids) | |
| antibody | Anti-DRP1 (mouse monoclonal) | Abcam | Abcam:ab56788 | (1:2000) |
| antibody | Anti-MFF (rabbit polyclonal) | SIGMA-ALDRICH | SIGMA:HPA010968 | (1:50) |
| antibody | Anti-MFF (rabbit polyclonal) | Protein Tech Group, Inc. | Proteintech: 17090–1-AP | (1:2000) |
| antibody | Anti-α-Tubulin | SIGMA-ALDRICH | SIGMA:T5168 | (1:2000) |
| recombinant DNA reagent | GFP-DRP1 (plasmid) | PMID:26101352 | ||
| recombinant DNA reagent | Mid49-Cherry (plasmid) | PMID:26101352 | ||
| recombinant DNA reagent | mCherry-DRP1 (plasmid) | PMID:21885730 | ||
| recombinant DNA reagent | mCherry-Lifeact (plasmid) | PMID:22980331 | ||
| recombinant DNA reagent | ATL1-K80A; CLIMP-63 (plasmid) | other | Robin Klemm (Zurich) | |
| recombinant DNA reagent | Cyto-ATL2 (plasmid) | PMID:28826471 | ||
| recombinant DNA reagent | pLVX-puro-GFP-Lifeact; pLVX-puro-RFP-Lifeact; pPax2; pMD2.G (plasmid) | other | Michael Way (London) | |
| recombinant DNA reagent | pLVX-GFP-Lifeact; pLVX-RFP-Lifeact (plasmid) | this paper | puromycin-resistant cassette deleted. | |
| recombinant DNA reagent | pLVX-mtBFP (plasmid) | this paper | Progentiors: PCR, mtBFP from pcDNA3.1-mtBFP (PMID:26259702); Vector pLVX | |
| recombinant DNA reagent | pLVX-Puro-MFF (plasmid) | this paper | Progenitors: PCR, GFP-MFF (Addgene:49153); Vector pLVX-puro | |
| recombinant DNA reagent | pLVX-mCherry-Fis1TM (plasmid) | this paper | Progenitors: PCR, Fis1 transmembrane domain from pBK416 (PMID:28864540); Vector pLVX | |
| recombinant DNA reagent | shCtrl (plasmid) | SIGMA-ALDRICH | SIGMA:SHC201 | |
| recombinant DNA reagent | shDRP1 (plasmid) | SIGMA-ALDRICH | SIGMA: TRCN0000318425 | |
| recombinant DNA reagent | pX330-DRP1e × 2 (plasmid) | this paper | pX330-DRP1e × 2 was generated by cloning the following annealed oligodeoxynucleotides into the pX330 vector (5’-caccGTGACAATTCCAGTACC TCT-3’, 5’-aaacAGAGGTACTGGAATT GTCAC-3’;) | |
| recombinant DNA reagent | pX330-DRP1e × 6 (plasmid) | this paper | pX330-DRP1e × 6 was generated by cloning the following annealed oligodeoxynucleotides into the pX330 vector (5’- caccGAGACCTCTCATTC TGCAAC-3’, 5’- aaacGTTGCAGAATGAG AGGTCTC-3’) | |
| sequence-based reagent | siDRP1 #1 | PMID:15286177 | siRNA 5'-UCCGUGAUGAGUAUG CUUUdTdT-3' | |
| sequence-based reagent | siDRP1 #2 | PMID:21186368 | siRNA 5′-CTGGAGAGGAATGCTGAAA-3′ | |
| sequence-based reagent | siMFF | this paper | siRNA 5′-CUGAGCAGUUCUGCA GUAACAdTdT-3′ | |
| sequence-based reagent | siINF2-CAAX | PMID:23349293 | siRNA 5′-ACAAAGAAACTGTGTGTGA-3′ | |
| software, algorithm | ndsafir | PMID:19900849 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30292.034






