A versatile genetic tool for post-translational control of gene expression in Drosophila melanogaster
Figures
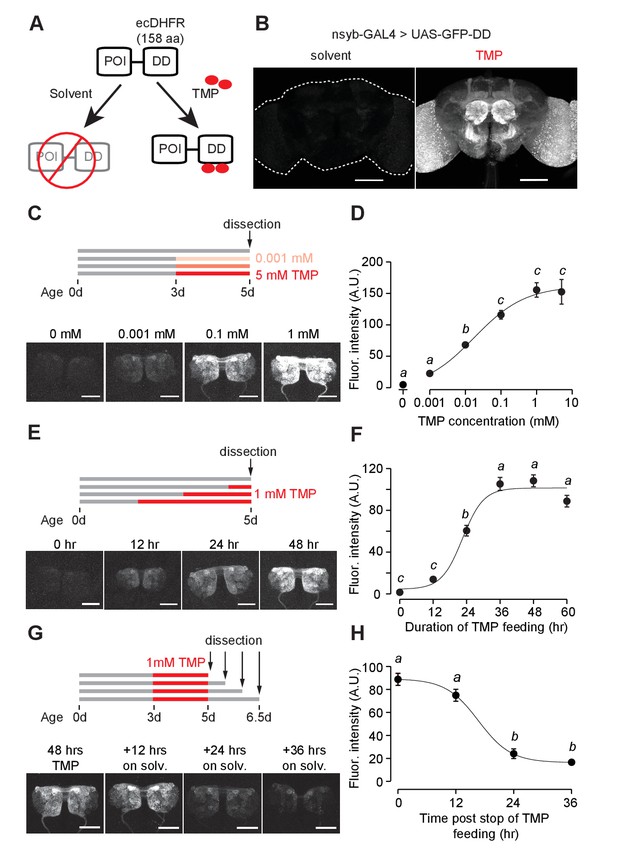
GFP-DD expression and degradation kinetics.
(A) Schematic of the destabilizing domain (DD) system. ecDHFR = E. coli. dihydrofolate reductase, POI = protein of interest, TMP = trimethoprim. (B) TMP-dependent GFP expression in the adult brain. Flies were fed 1 mM TMP-containing food from embryo stage up to dissection. (C, D) Dose-dependent change in GFP-DD expression in the axonal terminals of olfactory sensory neurons. Orco-Gal4, UAS-GFP-DD flies were fed with TMP (0–5 mM) for 48 hr before dissection (n = 5–6, p<0.001, F = 41.37, one-way ANOVA with Tukey’s post-hoc test). (E, F) GFP-DD expression is dependent on duration of TMP feeding. All flies were fed with fly food containing 1 mM TMP (n = 5–6, p<0.001, F= 87.34, one-way ANOVA with Tukey’s post-hoc test). (G, H) GFP degradation kinetics. Flies were fed with 1 mM TMP for 48 hr and then switched to standard fly food. GFP-DD expression in the antennal lobe was observed at 12 hr intervals following the switch. (n = 8–10, p<0.001, F = 71.43, one-way ANOVA with Tukey’s post-hoc test). Error bars indicate SEM. Significant differences between conditions (p<0.05) are denoted by different letters. Scale bar = 100 μm (B), 50 μm (C, E, G).
-
Figure 1—source data 1
Dosage dependence and kinetics of GFP-DD expression in response to TMP feeding.
- https://doi.org/10.7554/eLife.30327.010
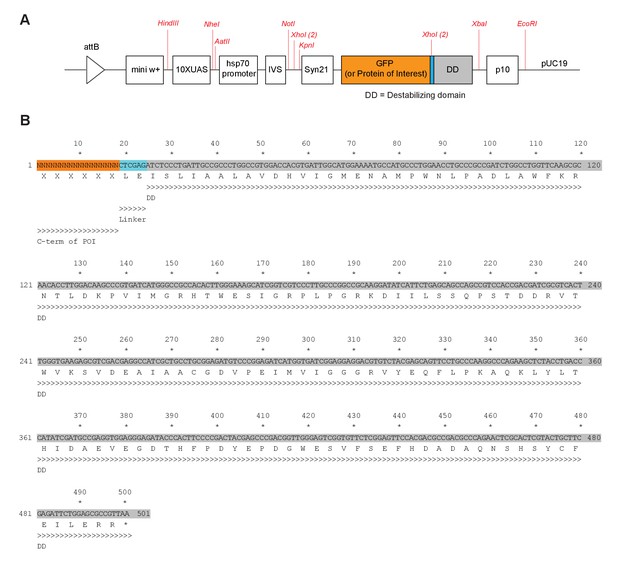
GFP-DD construct and sequence of the destablizing domain.
(A) Schematic of the 10XUAS-GFP-DD construct. Restriction sites are shown in red to facilitate cloning. All shown sites are unique apart from XhoI which has two cut sites (both shown). (B) DNA and protein sequence of the destabilizing domain.
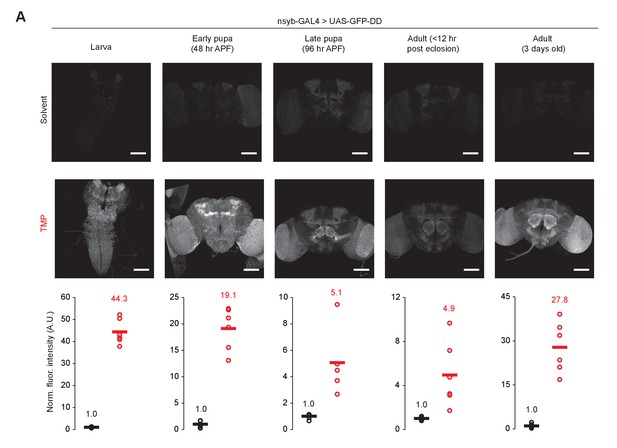
TMP-dependent GFP expression in the whole brain across developmental stages.
Panels from left to right: Larva, Early pupa (48 hr APF), Late pupa (96 hr APF), Adult (<12 hr post eclosion), Adult (3 day old). Fluorescence intensity has been normalized to the mean fluorescence intensity from the brains of flies fed with the solvent. Scale bar = 100 μm. Differences in GFP expression between solvent and TMP fed flies are significant (unpaired t-test, two-tailed) across all developmental stages - Larva (p<0.001, t = 18.6, n = 6), Early pupa (p<0.001, t = 10.89, n = 5–6), Late pupa (p=0.025, t = 3.46, n = 5–6), Adult at eclosion (p=0.022, t = 3.25, n = 6), 3 day old adult (p<0.001, t = 7.55, n = 6).
-
Figure 1—figure supplement 2—source data 1
Effect of TMP feeding on GFP-DD fluorescent intensity across developmental stages.
- https://doi.org/10.7554/eLife.30327.005
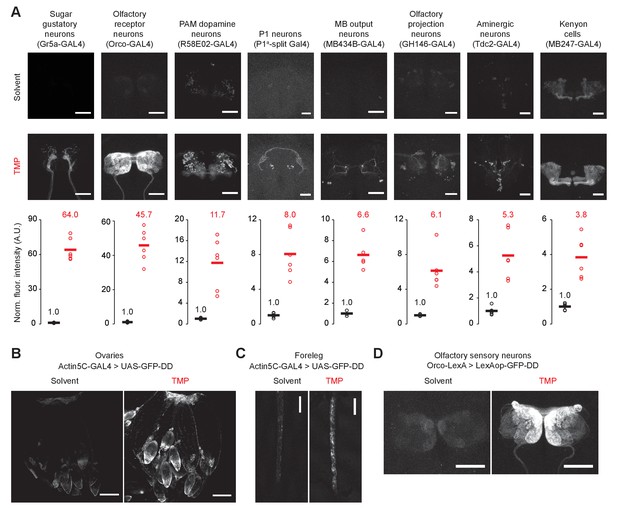
TMP-dependent GFP expression in different tissues.
(A) TMP-dependent GFP expression in different cell types in the adult brain. Fluorescence intensity has been normalized to the mean fluorescence intensity from the brains of flies fed with the solvent. Flies were fed with solvent or TMP-containing food from embryo stage to five days post-eclosion. Differences in GFP expression between solvent and TMP-fed flies are significant for all cell-types shown (n = 4–6, p<0.003, t > 5.3, unpaired t-test, two-tailed, for all cell types). (B, C) TMP-dependent GFP expression in ovaries (B) and the foreleg (C). (D) TMP-dependent GFP expression in olfactory sensory neurons driven by the LexA/LexAop system. Scale bar = 50 μm (A, D), 100 μm (B), 150 μm (C).
-
Figure 1—figure supplement 3—source data 1
TMP-dependent GFP expression in different tissues.
- https://doi.org/10.7554/eLife.30327.007
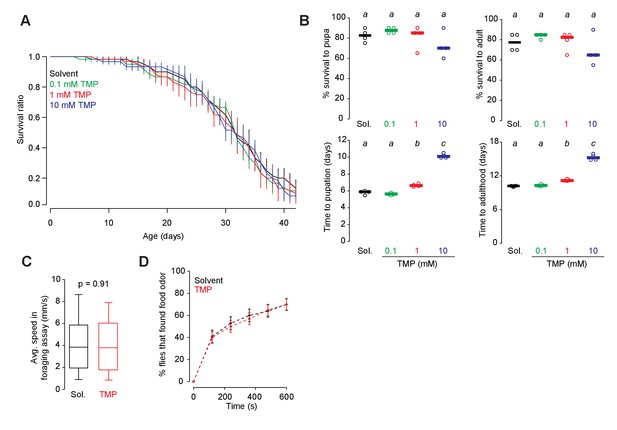
Effect of TMP on survival and behavior.
(A) Survival of wild-type flies fed with 0–10 mM TMP from eclosion to death. 15 flies were placed in each vial, four vials per condition. Error bars indicate SEM. (B) Survival (top) and developmental timing (bottom) of flies raised on 0–10 mM TMP. 20 eggs were placed in each vial, four vials per condition, one-way ANOVA with Tukey’s post-hoc test. Significant differences between conditions (p<0.05) are denoted by different letters. Survival to pupa (n = 4, p=0.186, F = 1.88), survival to adult (n = 4, p=0.245, F = 1.58), time to pupation (n = 4, p<0.0001, F = 389.04), and time to adulthood (n = 4, p<0.0001, F = 251.73) (C) Walking speed of wild-type (CS) flies fed with 1 mM TMP or solvent for 48 hr prior to assay (n = 159–161, unpaired t-test, two-tailed). Bar indicates median. Whiskers indicate 90% percentile. (D) Percentage of flies reaching the food odor is plotted against time. Food odor: 1% apple cider vinegar. Error bars indicate SEM.
-
Figure 1—figure supplement 4—source data 1
Effect of TMP on survival and behavior.
- https://doi.org/10.7554/eLife.30327.009
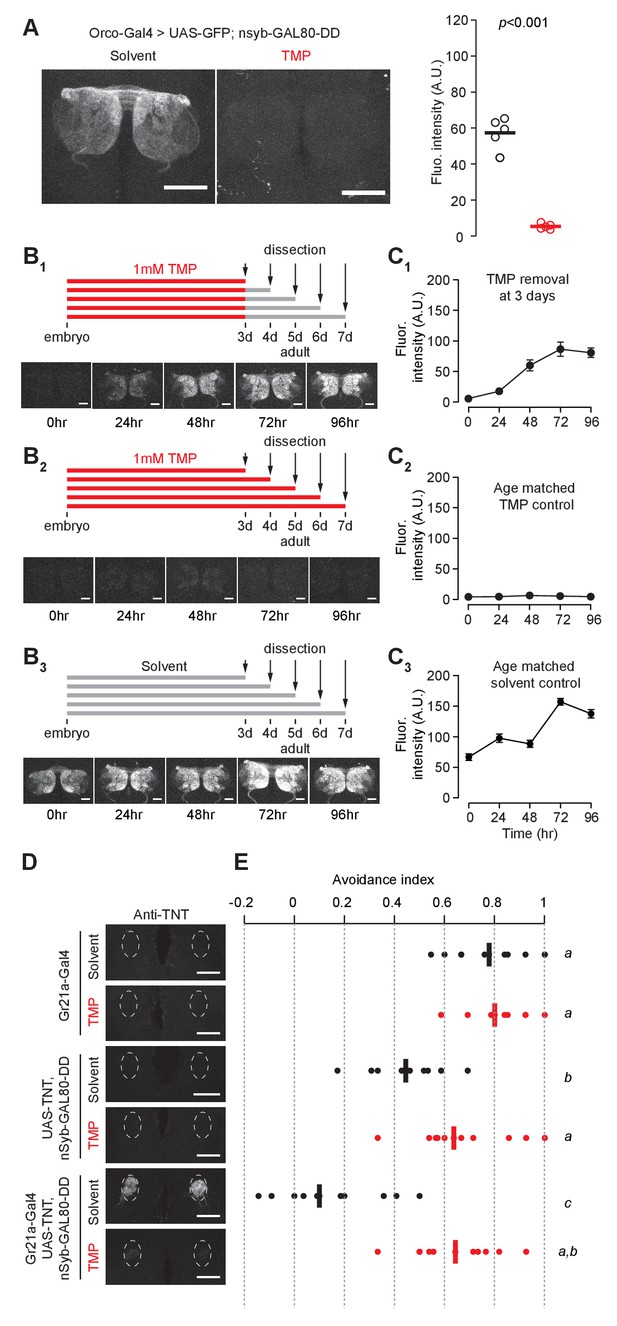
Chemically inducible control of GAL4-dependent expression by destabilized GAL80 (nsynaptobrevin-GAL80-DD).
(A) GAL4-driven GFP expression in olfactory sensory neurons can be suppressed by destabilized GAL80 (nsyb-GAL80-DD) in a TMP-dependent manner (n = 5, unpaired t-test, two-tailed, t = 13.25). (B, C) GAL80-DD can be used to temporally control GFP expression. (B1, C1) Orco-Gal4, UAS-GFP, nsyb-GAL80-DD flies were fed with food containing 1 mM TMP up to 3 days post-eclosion, following which flies were switched to standard fly food and dissected for quantification. GFP expression was compared to flies fed with 1 mM TMP throughout (B2, C2) or solvent throughout (B3, C3) (n = 4–5). 0 hr time point in C1 and C2 represent the same sample. (D) Tetanus toxin expression in the V glomerulus of flies fed with 1 mM TMP or solvent. (E) CO2 avoidance index of flies fed with 1 mM TMP or solvent. One arm of the T-maze contained 0.28% (v/v) CO2 and the other arm had air. GAL80-DD can restore CO2 aversion by suppressing TNT expression in the presence of TMP. n = 11 per condition, two-way ANOVA indicated a significant interaction between feeding condition and genotype, F = 23.66, p<0.001. Significant differences between conditions (p<0.05) are denoted by different letters (Tukey's post-hoc test). All flies were between 4–7 days old. Error bars indicate SEM. Scale bar = 50 μm (A), 25 μm (B,D).
-
Figure 2—source data 1
Chemically inducible control of GAL4-dependent expression by GAL80-DD.
- https://doi.org/10.7554/eLife.30327.014
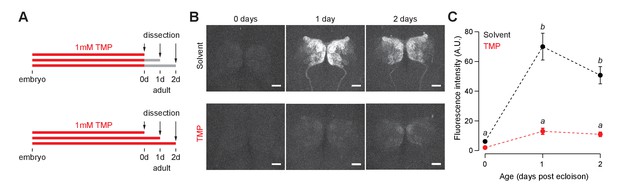
Functional characterization of GAL80-DD in early adults.
(A) Orco-Gal4, UAS-GFP, nsyb-GAL80-DD flies were fed with food containing 1 mM TMP up to eclosion, following which flies were switched to standard food or maintained on 1 mM TMP. (B,C) After switching to standard fly food, GFP expression is visible in the antennal lobe starting at 1 day post-eclosion. No GFP is observed in either TMP or solvent-fed flies right after eclosion (<8 hr adults). n = 5–6 per condition, two-way ANOVA indicated a significant interaction between feeding condition and age, F = 16.22, p<0.001. Significant differences between conditions (p<0.05) are denoted by different letters (Tukey's post-hoc test). Error bars indicate SEM. Scale bar = 25 μm.
-
Figure 2—figure supplement 1—source data 1
Functional characterization of GAL80-DD in early adults.
- https://doi.org/10.7554/eLife.30327.013
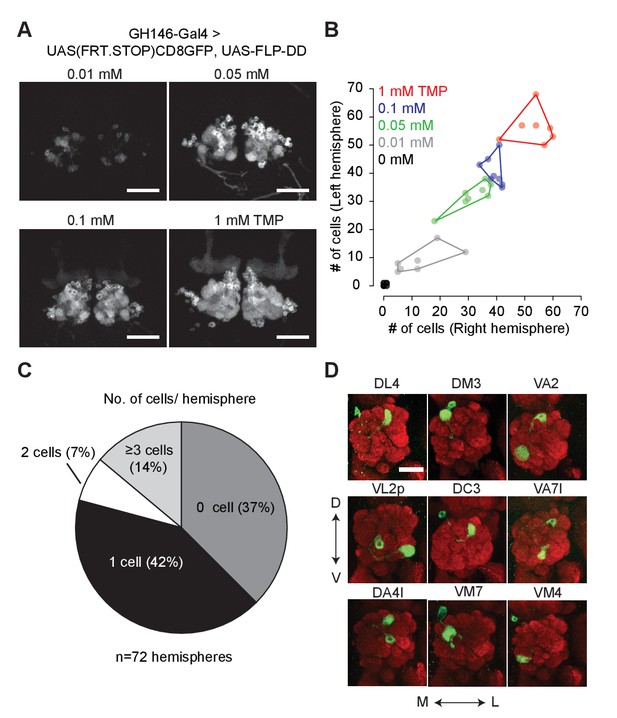
Chemical control of recombination frequency by destabilized flippase (10XUAS-FLP-DD).
(A) GFP expression in a sub-population of olfactory projection neurons following excision of the STOP cassette by FLP-DD. Scale bar = 50 μm. (B) Number of GFP-positive projection neurons can be controlled by varying TMP dosage. The number of GFP-labeled cells within a sample is similar across both brain hemispheres. Each point represents number of cells in one brain. (C) Pie chart indicating the number of labeled projection neurons for flies fed with solvent. 42% of all hemispheres had a single GFP-labeled cell. (D) Examples of labeled single projection neurons (D - dorsal, V - ventral, M - medial, L - lateral). Red = anti Bruchpilot nc82, Green = GFP. Scale bar = 25 μm.
-
Figure 3—source data 1
Chemical control of recombination frequency by FLP-DD.
- https://doi.org/10.7554/eLife.30327.016
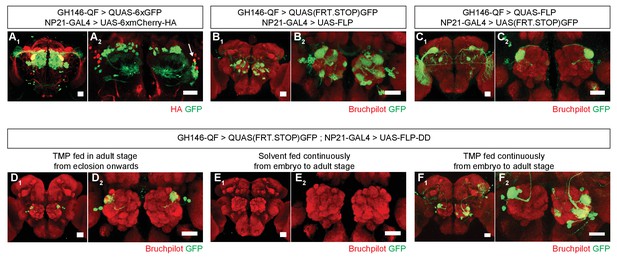
Refining intersection patterns by temporally limiting FLP-DD expression.
(A) Z-stack projections showing expression patterns of GH146-QF (green) and NP21-GAL4 (red). Both transgenic lines overlap in a single population of DA1 lPNs (arrow in A2). Between one to three overlapping neurons can be observed across all samples. Antenna was ablated from the brain sample shown in A2 to visualize projection neurons in the absence of sensory neuron axon terminals in the antennal lobe. (B–C) Intersection using constitutively expressed flippase generates expanded patterns with additional expression in other olfactory (B) or visual (C) projection neurons. (D) Temporally limiting FLP-DD expression by feeding 1 mM TMP exclusively in adult stage results in GFP expression only in DA1-lPNs. (E) No GFP expression is observed in the absence of TMP. (F) GFP expression in additional olfactory projection neurons can be observed using FLP-DD if TMP is fed continuously. Scale bar = 25 μm.
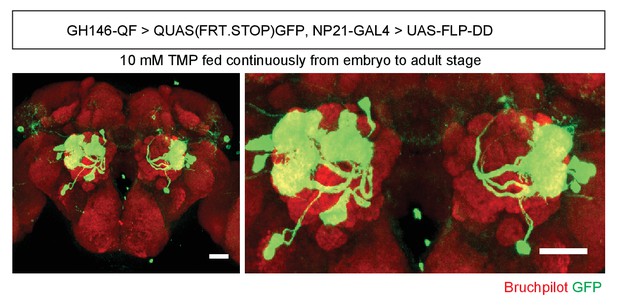
Intersection pattern between GH146-QF and NP21-GAL4 in flies fed with 10 mM TMP continuously.
Scale bar = 25 μm.
Tables
| Reagent type | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| genetic reagent (fly line) | nsyb-GAL4 | (Riabinina et al., 2015) | RRID:BDSC_51941 | |
| genetic reagent (fly line) | Orco-GAL4 | (Kreher et al., 2005) | RRID:BDSC_23292 | |
| genetic reagent (fly line) | UAS-GFP | |||
| genetic reagent (fly line) | GH146-GAL4 | (Stocker et al., 1997) | RRID:BDSC_30026 | |
| genetic reagent (fly line) | Gr5a-GAL4 | (Thorne et al., 2004) | RRID:BDSC_57591 | |
| genetic reagent (fly line) | R58E02-GAL4 | (Liu et al., 2012) | RRID:BDSC_41347 | |
| genetic reagent (fly line) | P1a-split GAL4 | (Hoopfer et al., 2015) | ||
| genetic reagent (fly line) | MB434B-split GAL4 | (Aso et al., 2014) | RRID:BDSC_68325 | |
| genetic reagent (fly line) | Tdc2-GAL4 | (Cole et al., 2005) | RRID:BDSC_9313 | |
| genetic reagent (fly line) | MB247-GAL4 | (Zars et al., 2000) | ||
| genetic reagent (fly line) | UAS-(FRT.STOP)mCD8-GFP | (Potter et al., 2010) | RRID:BDSC_30032 | |
| genetic reagent (fly line) | UAS-(FRT.STOP)GFP.myr | RRID:BDSC_55810 | ||
| genetic reagent (fly line) | UAS-6XmCherry-HA | (Shearin et al., 2014) | RRID:BDSC_52267 | |
| genetic reagent (fly line) | QUAS-6xGFP | (Shearin et al., 2014) | RRID:BDSC_52264 | |
| genetic reagent (fly line) | 20XUAS-FLPD5 | (Nern et al., 2011) | RRID:BDSC_55805 | |
| genetic reagent (fly line) | GH146-QF | (Potter et al., 2010) | RRID:BDSC_30014 | |
| genetic reagent (fly line) | QUAS(FRT.STOP)GFP | (Potter et al., 2010) | RRID:BDSC_30134 | |
| genetic reagent (fly line) | NP21-GAL4 | (Hayashi et al., 2002) | RRID:BDSC_30027 | |
| genetic reagent (fly line) | Actin5C-GAL4 | RRID:BDSC_4414 | ||
| genetic reagent (fly line) | Orco-LexAVP16 | (Lai et al., 2008) | ||
| genetic reagent (fly line) | Gr21-GAL4 | (Scott et al., 2001) | ||
| genetic reagent (fly line) | UAS-TNT | (Sweeney et al., 1995) | ||
| genetic reagent (fly line) | 10XUAS-GFP-DD | this study | see Materials and methods | |
| genetic reagent (fly line) | 10XUAS-FLP-DD | this study | see Materials and methods | |
| genetic reagent (fly line) | nsyb-GAL80-DD | this study | see Materials and methods | |
| genetic reagent (fly line) | 13XLexAop-GFP-DD | this study | see Materials and methods | |
| antibody | Rabbit anti-GFP | Invitrogen | A-11122, RRID:AB_221569 | 1:200 |
| antibody | mouse anti-bruchpilot nc82 | DSHB | RRID:AB_2314866 | 1:50 |
| antibody | mouse anti-HA | Biolegend | 901501, RRID:AB_2565006 | 1:500 |
| antibody | rabbit anti-TeTx | Statens Serum Institut | POL 016 | 1:1000 |
| antibody | Alexa Fluor 488 anti-rabbit immunoglobulin G | Invitrogen | A-31628, AB_143165 | 1:100 |
| antibody | Alexa Fluor 647 anti-mouse immunoglobulin G | Invitrogen | A-21235, AB_2535804 | 1:100 |
| recombinant DNA reagent (plasmids) | pJFRC81 | Pfeiffer et al., 2012 | Addgene_ 36432 | |
| recombinant DNA reagent (plasmids) | pJFRC95 | Pfeiffer et al., 2012 | ||
| recombinant DNA reagent (plasmids) | nsyb-GAL4-hsp70 | Riabinina et al., 2015 | Addgene_46107 | |
| recombinant DNA reagent (plasmids) | pCaSpeR-DEST5 | DGRC | 1031 | |
| recombinant DNA reagent (plasmids) | pAC-GAL80 | Addgene | 24346 | |
| recombinant DNA reagent (plasmids) | 10XUAS-GFP-DD | this study | see Materials and methods | |
| recombinant DNA reagent (plasmids) | 10XUAS-FLP-DD | this study | see Materials and methods | |
| recombinant DNA reagent (plasmids) | nsyb-GAL80-DD | this study | see Materials and methods | |
| recombinant DNA reagent (plasmids) | 13XLexAop-GFP-DD | this study | see Materials and methods | |
| chemical compound, drug | Trimethoprim | Teknova Inc., CA | T1205 | |
| software, algorithm | Igor Pro V6.0 | Wavemetrics, Inc. | RRID:SCR_000325 | |
| software, algorithm | Foraging assay behavior quantification | (Zaninovich et al., 2013) | Code available from (Zaninovich et al., 2013) | |
| other | Focusclear mounting reagent | Cedarlane Labs | FC-101 |
Additional files
-
Supplementary file 1
Genotypes and feeding conditions listed by figure and experiment.
- https://doi.org/10.7554/eLife.30327.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30327.020





