The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes
Figures
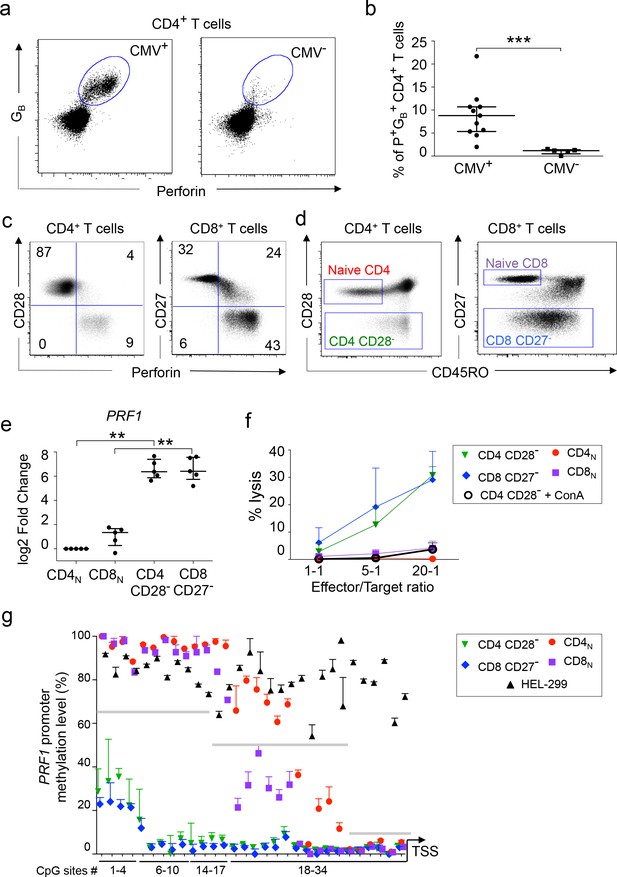
Phenotype and function of in vivo differentiated perforin+human CD4 T cells.
(a–b). The expression of perforin (P) and granzyme B (GB) was analyzed by flow cytometry in total CD4 T cells of CMV seropositive (CMV+) and seronegative (CMV-) healthy adults. (a) Representative dot plots of log10 fluorescence. (b) Proportions (median ±interquartile range) of P+GB+ CD4 T cells in 11 CMV+ and 5 CMV- subjects. (c) Perforin expression in CD28- CD4 T cells and CD27- CD8 T cells of CMV+ subjects Numbers indicate cell proportions in each quadrant. (d) Sorting strategy of naive and cytotoxic T cells according to expression of CD45RO, CD28 and CD27. Representative dot plot of log10 fluorescence. (e) The expression of PRF1 mRNA was measured by qPCR in purified T cell subsets of 5 CMV+ subjects. Results are median ± interquartile range of log2 fold change as compared to naive CD4 T cells. **:p<0,01 and ***:p<0,01. (f) The cytolytic activity of purified T cell subsets against anti-CD3-loaded target cells was assessed with or without pre-incubation with Concanamycin A (ConA). Data are mean ± SEM of three independent experiments on cells from different donors. (f) The methylation status of the PRF1 promoter was assessed in T cell subsets by bisulphite pyrosequencing. Data are median ± interquartile range of five donors for CD28-CD4 T cells and HEL-299 and of 9 donors for the other indicated subsets. Grey lines indicate three regions with distinct methylation profiles. See also Figure 1—figure supplement 1 and Source data file.
-
Figure 1—source data 1
Phenotype and function of in vivo differentiated perforin +human CD4 T cells.
Numerical data corresponding to the graphs of Figure 1 panels b, e, f and g.
- https://doi.org/10.7554/eLife.30496.004
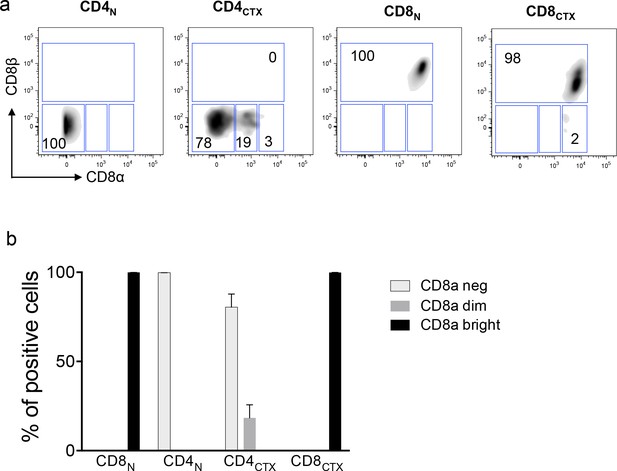
Expression of CD8 subunits by CD4CTX T cells.
The expression of CD8α and CD8β subunits by naive and cytotoxic CD4 and CD8 T cells was measured by flow cytometry. (a) Dot plot of a representative donor. (b) Percentage of cells expressing no (neg), intermediate (dim) or high (bright) levels of the CD8α subunit. Data are mean ±SEM% from three CMV-seropositive donors (b). The CD8β subunit was only expressed by CD8 T cells, whereas a proportion of CD4CTX cells expressed intermediate levels of CD8α.
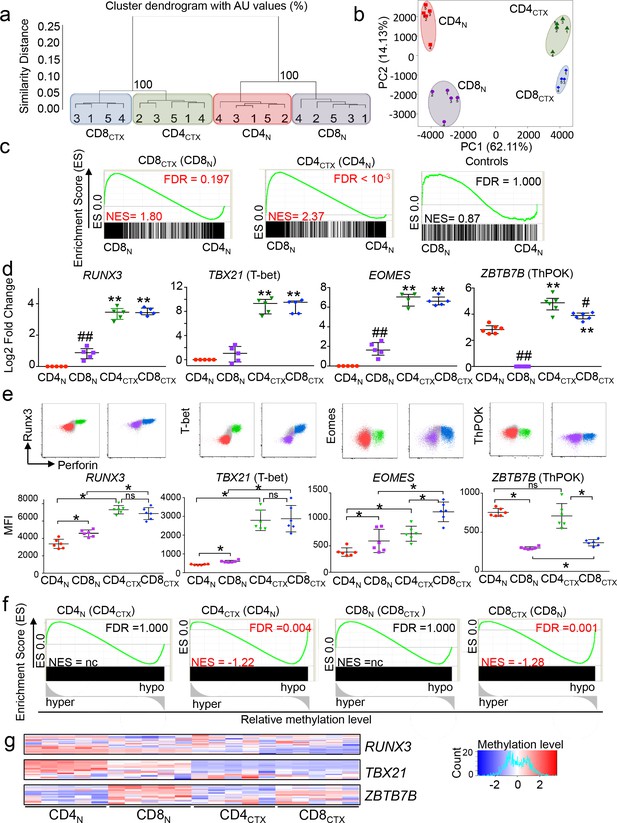
The transcriptional program of CD4CTX T cells is enriched in CD8 lineage genes without downregulation of ThPOK.
The transcriptome of T cell subsets from five CMV-seropositive donors (four for CD8CTX T cells) was analyzed by gene expression arrays. Log2 expression values of 14,372 probes with a variance >0.01 corresponding to 11,200 unique genes were submitted to unsupervised clustering (a) and principal component (b) analyses. (c) GSEA was used to test the enrichment of CD8CTX and CD4CTX T cell GeneSets (Supplementary file 1) in naive CD8 and naive CD4 T cell expression datasets. Genes showing no differential expression in CD8CTX and CD4CTX T cells were used as negative controls (n = 379). Bar codes show the ranking of the log2 fold change of gene expression values in naive CD8 versus naive CD4 T cells. Green lines represent enrichment profiles. False discovery rates (FDR) below 0.25 were considered significant and are indicated in red. NES: normalized enrichment score. (d) TF mRNA expression by T cell subsets purified from four to six donors, as indicated, was assessed by qPCR (upper panels). Results are expressed as median ±interquartile range of the log2 fold change as compared to naive CD4 or naive CD8 T cells. #:p<0.05 and ##:p<0.01 as compared to CD4 T cell counterparts; *:p<0.05 and **:p<0.01 as compared to naive counterparts. (e) TF protein expression was analyzed in T cell subsets by flow cytometry Upper panels: co-expression with perforin from one representative subject. Gated populations include naive CD4 (red), CD4CTX (green), naive CD8 (purple) and CD8CTX (blue) T cells. Lower panels: individual median intensity of fluorescence (MFI) of 5 CMV+ subjects. *:p<0.05. NS: not significant. Naive and cytotoxic T cell subsets were gated using the markers and strategy illustrated in Figure 1d. (f) GSEA was used to determine the correlation between gene expression and DNA methylation for each indicated T cell subset. Graphs show the enrichment of indicated GeneSets (Supplementary file 1) in genes that were either hypo or hypermethylated at the level of the promoter in the corresponding T cell subset. Black rectangles represent saturated bar codes of ranked delta beta values of 205,783 probes between the indicated subsets. (g) Heatmap of methylation beta values of RUNX3, TBX21 and ZBTB7B gene promoter in T cell subsets of five donors.. See also Figure 2—figure supplement 1 and Source data file.
-
Figure 2—source data 1
The transcriptional program of CD4CTX T cells is enriched in CD8 lineage genes without down regulation of ThPOK.
- https://doi.org/10.7554/eLife.30496.007
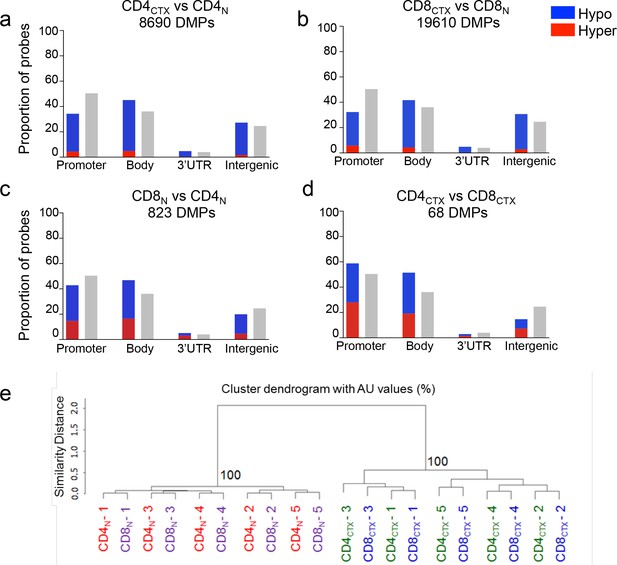
Whole genome methylation profiles of naive and cytotoxic CD4 and CD8 T cells.
Methylation profiles of 450,000 CpG sites were measured in T cell subsets with Infinium arrays in five CMV-seropositive healthy adults. The figure shows the proportions of hypomethylated (blue) and hypermethylated (red) probes identified in gene promoter, body, 3’UTR and intergenic regions following the comparison of CD4CTX and naive CD4 (CD4N) T cells (a), CD8CTX and naive CD8 (CD8N) T cells (b), CD8N and CD4N T cells (c) and CD4CTX and CD8CTX T cells (d). Grey bars indicate the proportions of probes at the different gene locations. The differentiation of naive in cytotoxic T cells was associated with the hypomethylation of large numbers of probes. Methylation values of 95,877 probes with a variance >0.01 were submitted to unsupervised clustering analysis (e).
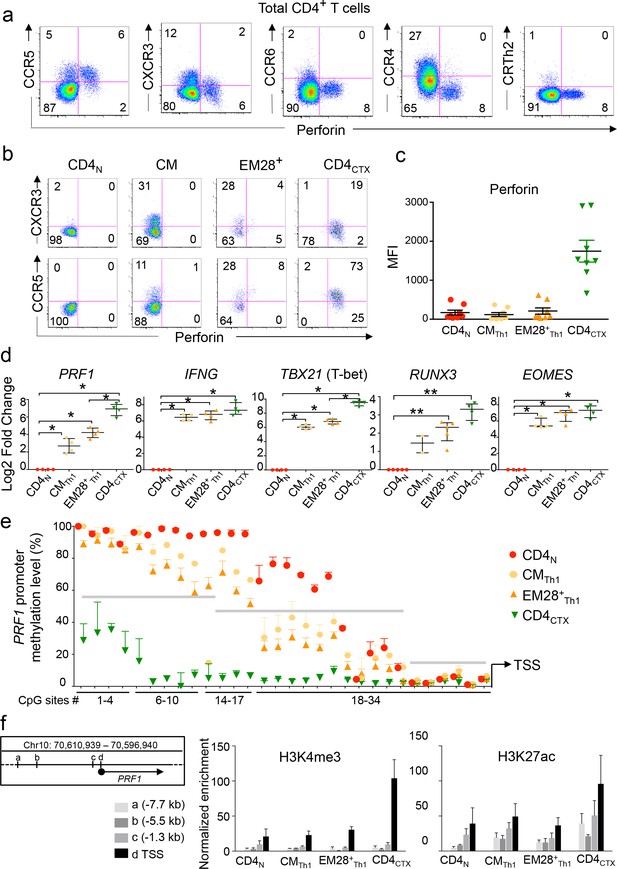
Differentiation of CD4CTX T cells within the Th1 lineage.
(a–b). Expression of perforin and chemokine receptors was assessed by flow cytometry in total CD4 T cells (a) central memory (CM), effector memory (EM) CD28+ and CD4CTX T cells (b). Log10 fluorescence of one representative donor out of 9. (c) Median intensity of fluorescence (MFI) of perforin expression in T cell subsets from 7 CMV+ subjects. (d) mRNA expression of indicated genes was assessed by qPCR in T cell subsets purified from two to five donors, as indicated. CMTh1 and EM28+Th1 were CCR5+CCR6- as illustrated in Supplementary Figure 3. Results are median ±interquartile range of the log2 fold change as compared to naive CD4 T cells. *:p<0.05 and **:p<0.01. (e) Methylation status of PRF1 promoter was assessed by bisulphite pyrosequencing in indicated purified T cell subsets. Results are presented as median ±interquartile range of two to five donors depending on the CpG site, as detailed in Supplementary file 2a. Grey lines indicate three regions with distinct methylation profiles. (e) Histone modifications at indicated regions of the PRF1 locus were studied by ChIP-qPCR in purified CD4 T cell subsets of three donors. Results are % of input after normalization for the enrichment in pan H3. Letters refer to indicated distances from the transcription start site (TSS) (left panel). See also Figure 3—figure supplements 1 and 2 and Source data file.
-
Figure 3—source data 1
Differentiation of CD4CTX T cells within the Th1 lineage.
Numerical data corresponding to the graphs of Figure 3 panels c, d and f.
- https://doi.org/10.7554/eLife.30496.011
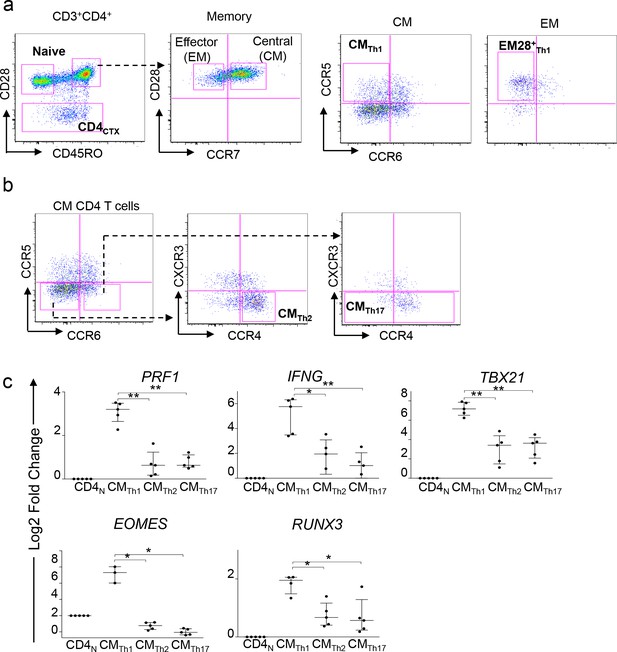
Sorting strategy of memory CD4 T cell subsets and expression of candidate genes.
(a) Sorting strategy of naive (CD3+CD4+CD45RO-CD28+), central memory Th1 (CMTh1) (CD3+CD4+CD45RO+CD28+CCR7+CCR5+CCR6-), CD28+effector memory Th1 (EM28+Th1) (CD3+CD4+CD45RO+CD28+CCR7-CCR5+CCR6-) and CD4CTX (CD3+CD4+CD28-) T cells. (b) Sorting strategy of CMTh2 (CD3+CD4+CD45RO+CD28+CCR7+CCR4+CCR6-CCR5-CXCR3-) and CMTh17 (CD3+CD4+CD45RO+CD28+CCR7+CCR6+CCR5-CXCR3-) cells. (c) The expression of candidate gene mRNA was measured by qPCR in purified T cell subsets from three to five donors. Data are median ±interquartile range of log2 fold change as compared to naive CD4 (CD4N) T cells. *:p<0.05 and **:p<0.01. CMTh1 cells expressed higher levels of PRF1, IFNG, TBX21, EOMES and RUNX3 than the other CM subsets.
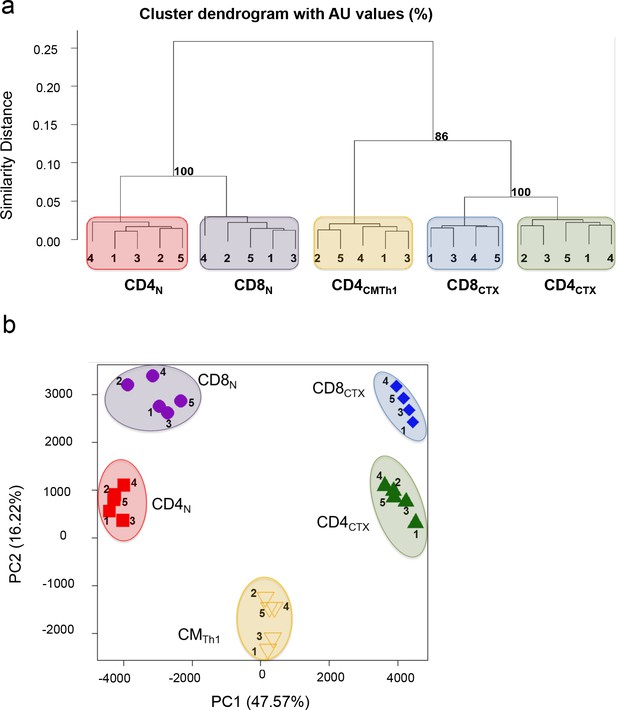
Transcriptional program underlying the expression of perforin in Th1 cells.
(a-b) The transcriptome of CD4 and CD8 T cell subsets purified from four to five CMV-seropositive donors was analysed by genome-wide mRNA expression arrays. Log2 expression values of 13,551 probes with a variance>0.01 corresponding to 10,699 unique genes were submitted to hierarchical clustering (a) and principal component (b) analyses.
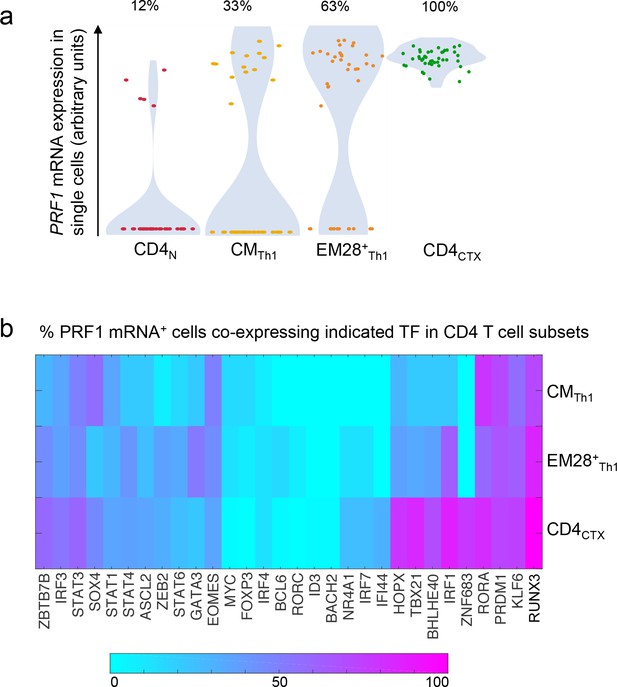
Proportion of PRF1 mRNA+cells and co-expression with transcription factors in the Th1 lineage.
The expression of PRF1 and transcription factors (TF) mRNA was analyzed in 43 single naive, central memory (CM) Th1, effector memory (EM) CD28+ Th1 and CD4CTX T cells from one donor. (a) PRF1 mRNA expression in single cells and proportions of PRF1+ cells in Th1 cell subsets. (b) Heat map of PRF1+ cells co-expressing individual TF in Th1 cell subsets. (calculated proportion: double-positive cells/perforin-positive cells). See also Figure 4—source data 1.
-
Figure 4—source data 1
Proportion of PRF1 mRNA +cells and co-expression with transcription factors in the Th1 lineage.
Single-cell expression values for each indicated genes (column) and each cell (lines).
- https://doi.org/10.7554/eLife.30496.013
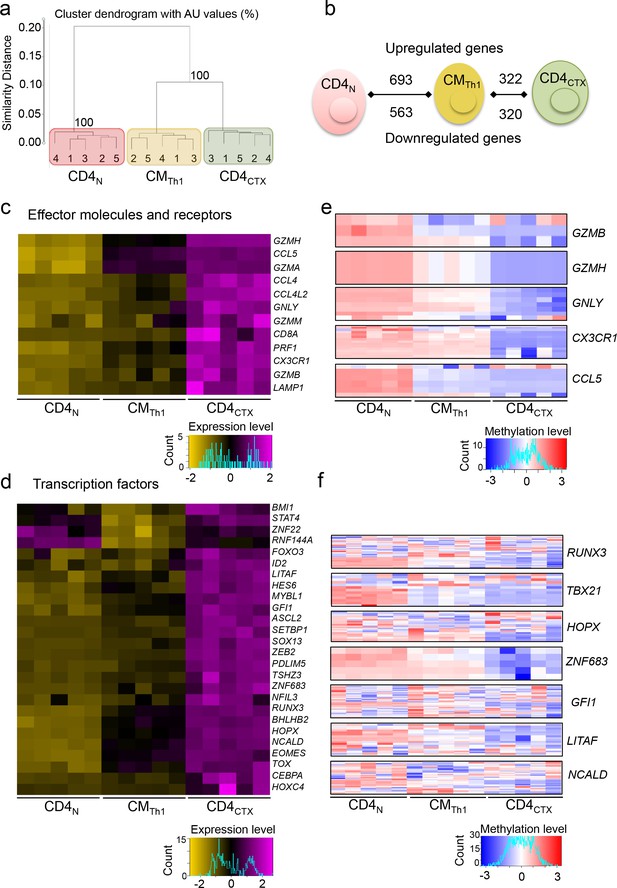
Transcriptional program underlying the expression of perforin in Th1 cells.
The transcriptome of CD4 T cell subsets from five donors was analyzed by gene expression arrays. (a) Log2 expression values of 13,551 probes with a variance >0.01 corresponding to 10,669 unique genes were submitted to unsupervised cluster analysis. See also Supplementary Figure 4. (b) Genes differentially expressed by naive CD4 (CD4N), central memory (CM) Th1 and CD4CTX T cells were identified. (c-d) Heatmaps of mean log2 expression values of all probes for each selected effector molecules and receptors (c) and transcription factors (d) upregulated in CD4CTX T cells as compared to CMTh1 cells. See also Figure 5—figure supplement 1. (e-f) Heatmaps of methylation beta values of all the probes located in the promoter region of effector molecules and receptors (e) and transcription factors (f) significantly hypomethylated in CD4CTX as compared to CMTh1 as assessed by a two-way ANOVA with Tukey’s multiple comparisons (p<0.01). See also Figure 5—figure supplement 2 and Source data file.
-
Figure 5—source data 1
Transcriptional program underlying the expression of perforin in Th1 cells.
Raw methylation data of all the probes included in the heatmap of Figure 5 panels e and f, including location on the gene.
- https://doi.org/10.7554/eLife.30496.017
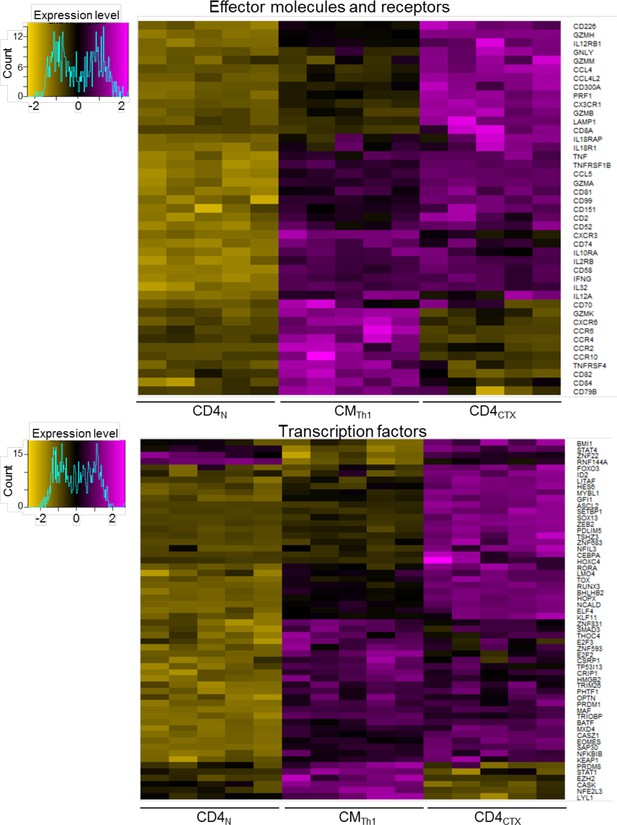
Transcriptional program of CMTh1 and CD4CTX T cells.
Heatmaps of mean log2 expression values of all probes for selected effector molecules and receptors (c) and transcription factors (d) up regulated in CMTh1 as compared to CD4N T cells or in CD4CTX T cells as compared to CMTh1 cells.
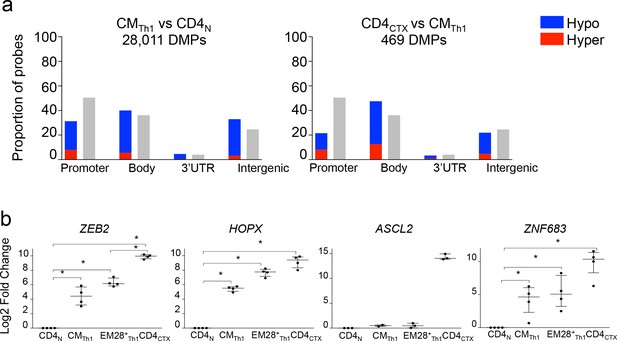
Epigenetic and transcriptional program underlying the expression of perforin in Th1 cells.
(a) Methylation profiles of 450,000 CpG sites were measured in T cell subsets with Infinium arrays. The figure shows the proportions of hypomethylated (blue) and hypermethylated (red) probes identified in gene promoter, body, 3’UTR and intergenic regions following the comparison of CMTh1 and naive CD4 (CD4N) T cells (left panel), CD4CTX and CMTh1 cells (right panel). Grey bars indicate the proportions of probes at the different gene locations. The differentiation of CMTh1 and CD4CTX T cells was associated with the hypomethylation of large numbers of probes. (b) The expression of candidate gene mRNA was measured by qPCR in purified T cell subsets from three to four donors. Data are median ± interquartile range of log2 fold change as compared to naive CD4 (CD4N) T cells. *:p<0.05. High levels of ZEB2, HOPX, and ZNF683 (Hobit) were detected in all memory CD4 T cell subsets whereas increased levels of ASCL2 were only detected in CD4CTX T cells.
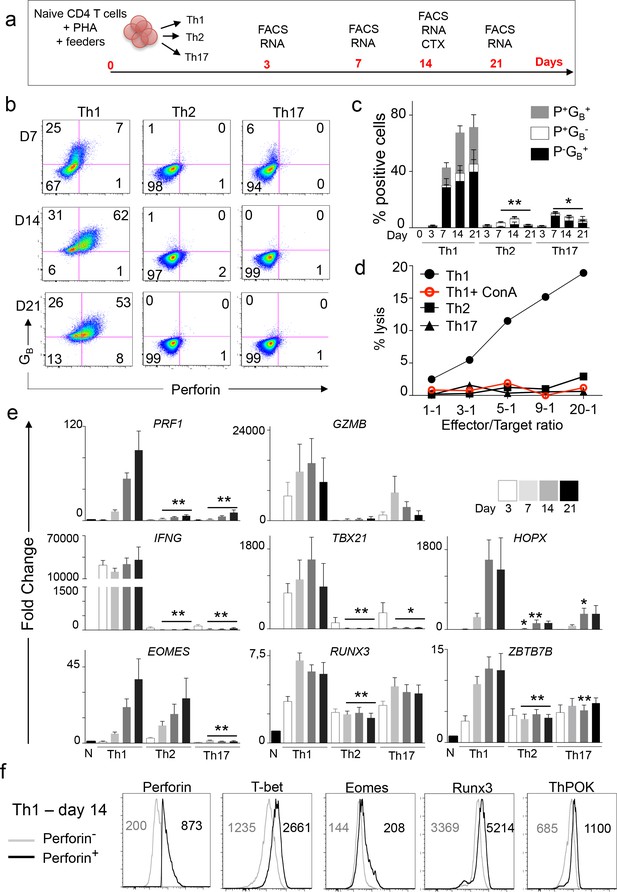
Naive CD4 T cells differentiate into CD4CTX in Th1 culture conditions in vitro.
(a) Naive CD4 T cells were stimulated polyclonally in the presence of Th1, Th2 or Th17 polarizing cytokines. Flow cytometry (FACS), gene expression (RNA) and cytotoxicity analyses were performed at the indicated time points. (b-c) Perforin (P) and GranzymeB (GB) expression was assessed by flow cytometry. (b) Dot plots (Log10 fluorescence of one representative out of six different donors. Numbers indicate % of cells in individual quadrants. (c) Mean ±SEM of % of perforin (P) and granzyme B (GB) positive cells from six independent experiments on different donors (only 4 for day 21 and 5 for day 3). (d) Cytotoxic activity of in vitro differentiated effector T cells against anti-CD3-loaded target cells was assessed at indicated effector/target ratios with or without pre-incubation with Concanamycin A. Figure shows one representative out of four experiments on different donors. (e) mRNA expression of indicated genes was quantified by qPCR. Results are mean ±SEM fold change as compared to naive CD4 T cells from six independent experiments on different donors (only 3 for day 3). *:p<0.05 and **:p<0.01 as compared to Th1 condition at the corresponding time point. (f) Expression of TF was measured by flow cytometry in perforinhigh and perforinlow Th1 cells after 14 days of in vitro stimulation. Histograms of one representative out of six experiments. Numbers are median fluorescence intensity (MFI) of six experiments (only four for ThPOK). See also Figure 6—figure supplements 1 and 2 and Source data file.
-
Figure 6—source data 1
Naive CD4 T cells differentiate into CD4CTX in Th1 culture conditions in vitro.
Numerical data corresponding to the graphs of Figure 6 panels c, d and e.
- https://doi.org/10.7554/eLife.30496.021
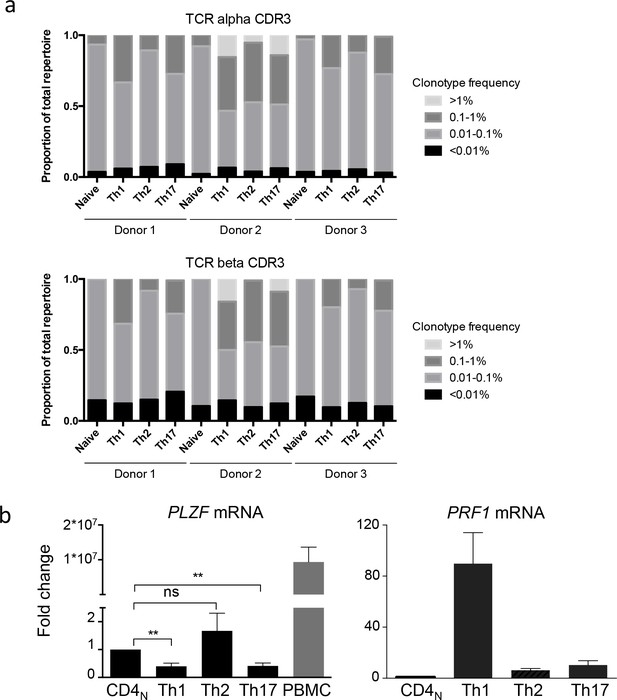
TCR repertoire of and PLZF expression by in vitro differentiated CD4 T cells.
(a) TCR alpha and beta repertoires of CD4 T cells was determined by CDR3 RACE PCR and sequencing at day 0 (Naive) and after 14 days of activation in the presence of polarizing cytokines. Data were obtained from three independent experiments on three different donors. Between 3000 and 6000 unique clonotypes were sequenced per experimental condition. Diverse repertoires including large (>1%) and medium (0.1–1%) clonotype expansions were detected in differentiated CD4 T cells. (b) PLZF and PRF1 mRNA expression by naive and differentiated CD4 T cells was measured by qPCR. Data are mean ±SEM fold change as compared to naive cells from seven independent experiments. **:p<0.01. PRF1 mRNA expression data are the same as those presented in Figure 6e. Th1 and Th17 cells had lower expression of PLZF mRNA. Together, these results indicate that CD4 T cells differentiated in the presence of Th1 cytokines have features of conventional and not innate-like lymphocytes.
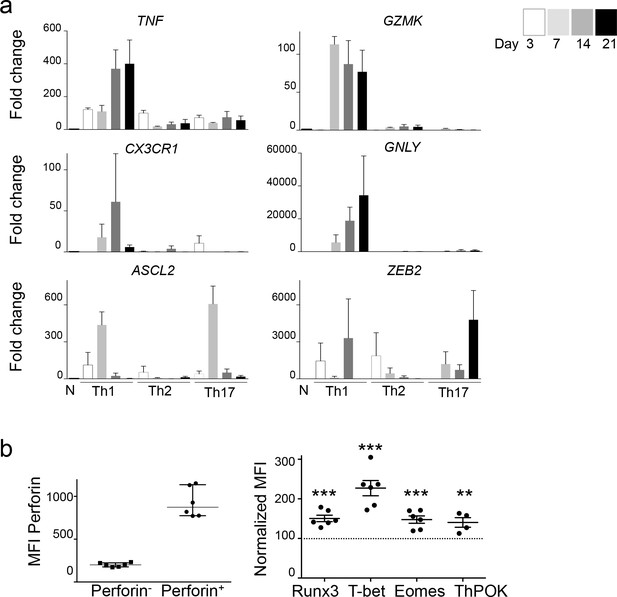
Expression of TF and effector molecules by in vitro differentiated CD4 T cells.
(a) The expression of mRNA coding for TF and effector molecules was measured by qPCR at various time points following the activation of naive CD4 T cells in the presence of Th1, Th2 or Th17 polarizing cytokines. Results are mean ±SEM fold change as compared to naive CD4 T cells from three to six independent experiments. Th1 cells expressed higher levels of TNF, GZMK, CX3CR1 than Th2 and Th17 cells, whereas ASCL2 and ZEB2 were expressed at high levels by more than one Th subset. (b) The expression of Runx3, T-bet, Eomes and ThPOK was measured by flow cytometry after 14 days of activation of naive CD4 T cells in the presence of Th1 polarizing cytokines. Results are median ±IC of MFI of perforin (left panel) and normalized MFI of transcription factors expression by perforin+ cells as compared to perforin- cells from four to six experiments. **:p<0.01, ***:p<0.001. Perforin+ cells expressed higher levels of Runx3, T-bet, Eomes and ThPOK than perforin- cells.
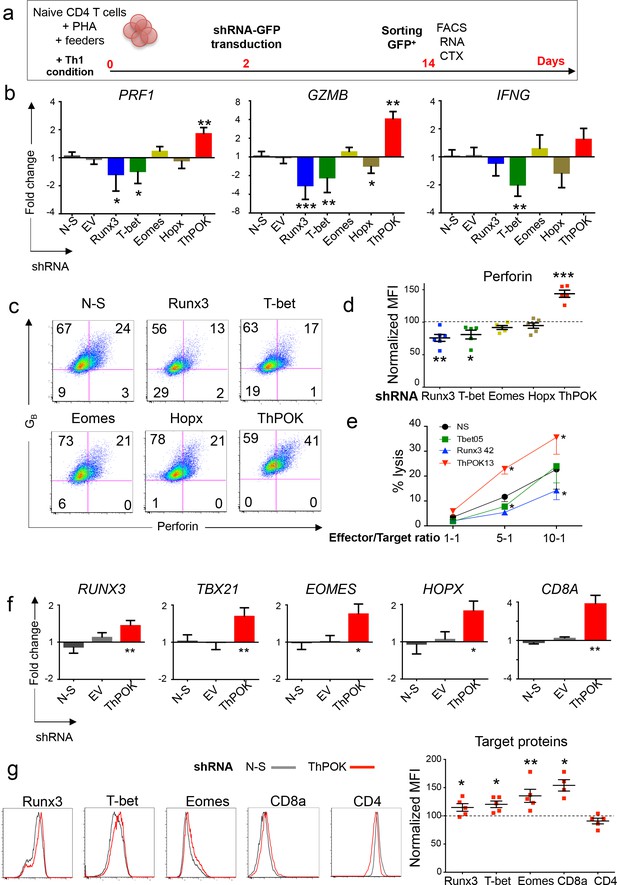
TF controlling the expression of perforin in CD4 T cells.
(a) Naive CD4 T cells were stimulated polyclonally in the presence of Th1 polarizing cytokines and were transduced on day 2 with shRNA-GFP. GFP+ cells were purified on day 14 for flow cytometry, mRNA expression and cytotoxicity analyses. (b) Expression of PRF1, GZMB and IFNG mRNA was quantified by qPCR in cells transduced with indicated targeting shRNA, non-silencing (N-S) shRNA or empty vector (EV). Data are mean ± SD log2 fold change as compared to controls (mean of N-S and EV), from seven biological replicates generated in four independent experiments on different donors. (c-d) Expression of perforin (P) and granzyme B (GB) was measured by flow cytometry in cells transduced with N-S or indicated gene targeting shRNA. (c) Representative dot plot (log10 fluorescence) of the proportions of double positive cells from seven independent experiments on different donors. Numbers indicate % of cells in individual quadrants. (d) Perforin expression following knockdown of indicated TF. Data are median fluorescence intensity (MFI) normalized for perforin MFI in cells transduced with N-S shRNA (dotted line, 100%). Experiments in which gene knockdown was below 10% were excluded from the analysis. (e) Cytotoxic activity of transduced cells against anti-CD3-loaded target cells. Data are mean ± SEM of five independent experiments on different donors. *:p<0.05 as compared to N-S shRNA. (f-g) The effect of ThPOK knockdown on the expression of indicated TF, CD8A and CD4 was studied by qPCR and flow cytometry. (f) Data are mean ± SD log2 fold change as compared to mean RNA expression in control shRNA (N-S and EV) from seven biological replicates generated in four independent experiments on different donors. *:p<0.05; **:p<0.01 as compared to control shRNA. (g) Histograms (Log10 fluorescence) from one representative donor. Data are MFI normalized for TF MFI in cells transduced with N-S shRNA (dotted line, 100%) of seven independent experiments on different donors. See also Figure 7—figure supplements 1 and 2 and Source data file.
-
Figure 7—source data 1
TF controlling the expression of perforin in CD4 T cells.
Numerical data corresponding to the graphs of Figure 7 panels b, d, e and g.
- https://doi.org/10.7554/eLife.30496.025
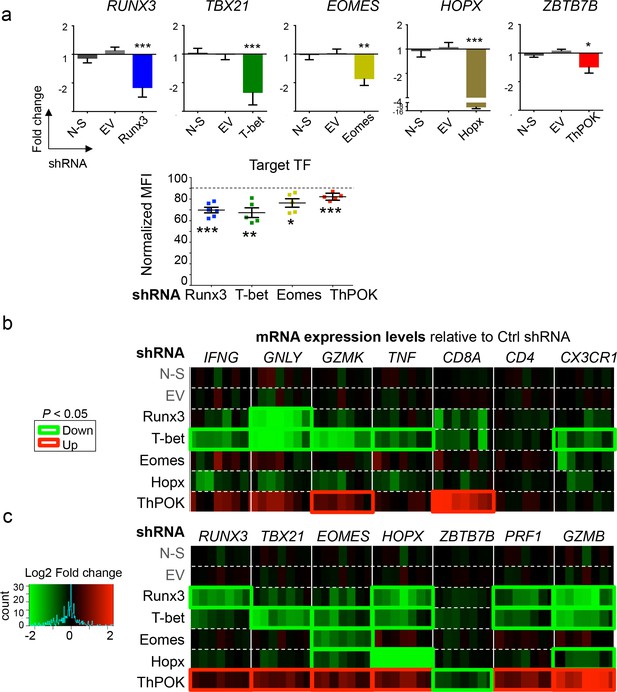
Knockdown of transcription factors (TF) expression by shRNA.
Specific shRNA were used to knockdown the expression of candidate TF by in vitro differentiated Th1 cells. (a) qPCR and flow cytometry analyses of the expression of TF, effector molecules and receptors. qPCR data are mean ± SD fold change as compared to controls (mean of non-silencing (N-S) shRNA and empty vector (EV)) of seven biological replicates from four independent experiments). Flow cytometry data are mean ± SEM of normalized median fluorescence intensity (MFI) as compared to N-S shRNA from seven independent experiments. Experiments in which less than 10% knockdown was achieved for Runx3, T-bet and Eomes were excluded from the analysis. *:p<0.05; **:p<0.01; ***:p<0.001. Results show effective knockdown of target TF. (b-c) Heatmaps of mRNA expression levels show that T-bet knockdown increases the expression of IFNG, GNLY, GZMK, TNF and CX3CR1; Runx3 knockdown increases the expression of GNLY; ThPOK knockdown increases the expression of GZMK, CD8A and all other target TF. In contrast, N-S shRNA and EV transduction did not affect expression of any analyzed mRNA.
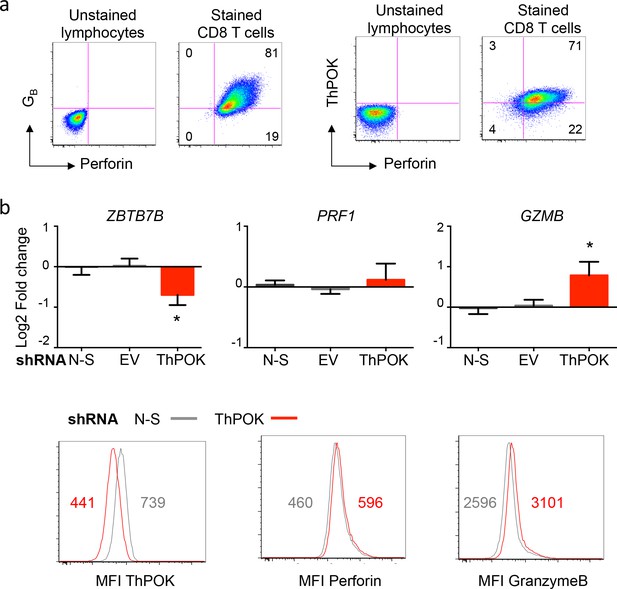
Influence of ThPOK on the differentiation of cytotoxic CD8 T cells.
Naive CD8 T cells were submitted to polyclonal stimulation with PHA in the presence of irradiated allogeneic feeder cells, IL-2, IL-15 and Th1 polarizing cytokines for 7 to 14 days. Perforin, granzyme B (GZMB) and ThPOK expression was measured by flow cytometry and by qPCR. (a) Differentiated CD8 T cells expressed perforin and ThPOK. Gates were placed using an unstained control and numbers indicate the % of cells in each quadrant. Similar results were obtained in three independent experiments. (b) Suppression of ThPOK expression in differentiated CD8 T cells was performed using specific shRNA as done in Figure 7. ThPOK knockdown, verified at mRNA and protein levels, did not significantly influence the expression of perforin but significantly increased the expression of granzyme B as compared to non-silencing (N-S) shRNA. Data are mean ± SD log2 fold change of five replicates from three independent experiments (upper panels) and one histogram with median fluorescence intensity (MFI) of two independent experiments. *:p<0.05. These results support the notion that ThPOK expression could limit the cytotoxic function of CD8 T cells, as observed in CD4 T cells.
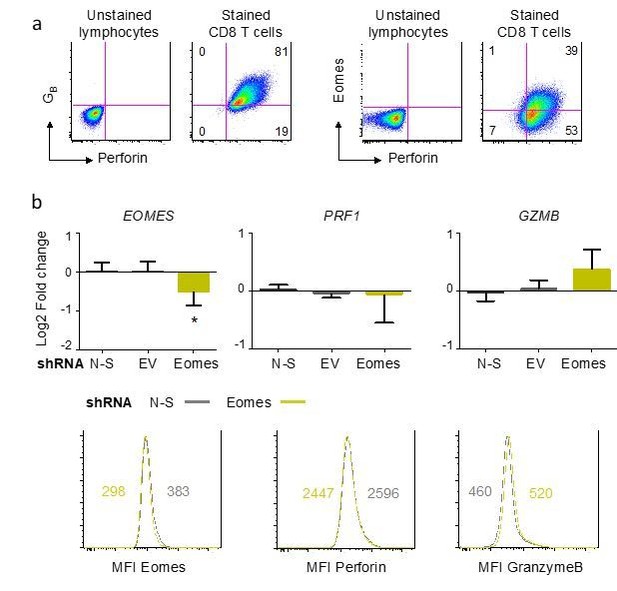
Influence of Eomes on the differentiation of cytotoxic CD8 T cells.
Naive CD8 T cells were submitted to polyclonal stimulation with PHA in the presence of irradiated allogeneic feeder cells, IL-2, IL-15 and Th1 polarizing cytokines for 7 to 14 days. Perforin, granzyme B (GZMB) and Eomes expression was measured by flow cytometry and by qPCR. a) Differentiated CD8 T cells expressed perforin and a fraction of them also expressed Eomes. Gates were placed using an unstained control and numbers indicate the% of cells in each quadrant. Comparable results were obtained in three independent experiments. b) Suppression of Eomes expression in differentiated CD8 T cells was performed using specific shRNA. Eomes knockdown, verified at mRNA and protein levels, did not significantly influence the expression of perforin or granzyme B as compared to non-silencing (N-S) shRNA. Data are mean +/- SD log2 Fold change of 5 replicates from 3 independent experiments (upper panels) and one histogram with median fluorescence intensity (MFI) of 2 independent experiments. *:p<0.05.
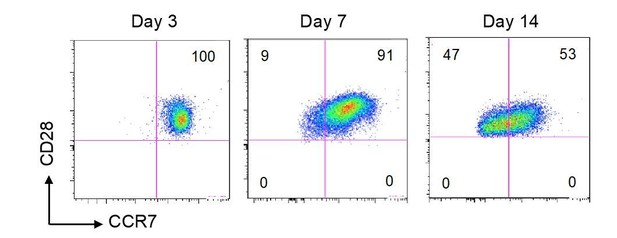
Stable CD28 expression upon 14 days in vitro stimulation.
Naive CD4 T cells were subjected to polyclonal stimulation as described in the manuscript (Figure 6). Results are flow cytometry plots of CD28 and CCR7 expression 3, 7 and 14 days after stimulation. Numbers indicate the% of cells in each quadrant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers |
|---|---|---|---|
| Cell line (mouse leukemia) | RAW 264.7 | ATCC Cat# TIB-71 | RRID:CVCL_0493 |
| Cell line (human kidney cell line) | HEK-293 human kidney cell line | ATCC Cat# CRL-1573, | RRID:CVCL_0045 |
| Cell line (human) | HEL 299 | ATCC Cat# CCL-137, | RRID:CVCL_2480 |
| Transfected constructs | References of all shRNAs are listed in Supplementary file 2f | ||
| Antibody-ChIP | References of used antibodies are indicated in the method section | ||
| Antibody-cytometry | References of all used cytometry antibodies, including company and clone are listed in Supplementary file 2b. | ||
| Sequence-based reagent | All sequences are listed in Supplementary files 2c, d and e | ||
| Chemical compound, drug | Concanamycin A | Sigma-Aldrich (Merck, Germany) | C 9705 |
| Other | PKH26 staining | Sigma-Aldrich (Merck, Germany) | Catalog numbers MINI26 and PKH26GL |
Additional files
-
Supplementary file 1
Lists of genes included in the 12 GeneSets obtained from CD4 versus CD8 T cell comparison (Figure 2) and naive CD4 T cell versus CMTh1 cell versus CD4CTX T cell comparison (Figure 4).
Genes expressed at higher level by the first listed subset as compared to the subset indicated between brackets were identified using the min/max method. Genes are ranked according to mean log2 fold change calculated using the Limma package in R.
- https://doi.org/10.7554/eLife.30496.026
-
Supplementary file 2
(a) Level of methylation at individual CpG sites in memory Th1 cell subsets in vivo.
The level of methylation was measured by pyrosequencing in memory Th1 cell subsets of two to nine CMV-seropositive healthy adults. Data are median percentage ± interquartile range and were compared with the Mann-Withney non-parametric test. † CMTh1 versus naive CD4 T cells. ‡ EM28+Th1 versus naive CD4 T cells. § CD4CTX versus naive CD4 T cells. ¶ EM28+Th1 versus CMTh1. || CD4CTX versus EM28+Th1. nd : not done, no statistical analysis was performed because of insufficient number of subjects (n = 2 or 3) ; ns : non significant ;*=p<0.05 ; **=p<0.01 ; ***=p<0.001 ; ****=p<0.0001. (b) Monoclonal antibodies used in flow cytometry experiments. (c). QPCR Oligonucleotide sequences (d) ChIP-PCR oligonucleotide sequences. (e) Single-cell qPCR oligonucleotide sequences. (f) References for the gene transfer plasmids used in transcription factor knockdown experiments.
- https://doi.org/10.7554/eLife.30496.027
-
Supplementary file 3
Method to analyze the correlation between gene expression and DNA methylation.
Gene Set Enrichment Analysis (GSEA) was used to analyze the enrichment of DNA methylation profiles in the GeneSets expressed by T cell subsets. Five Perl scripts were created to prepare the file required for GSEA.
- https://doi.org/10.7554/eLife.30496.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30496.029





