Genome-wide mapping of sister chromatid exchange events in single yeast cells using Strand-seq
Figures
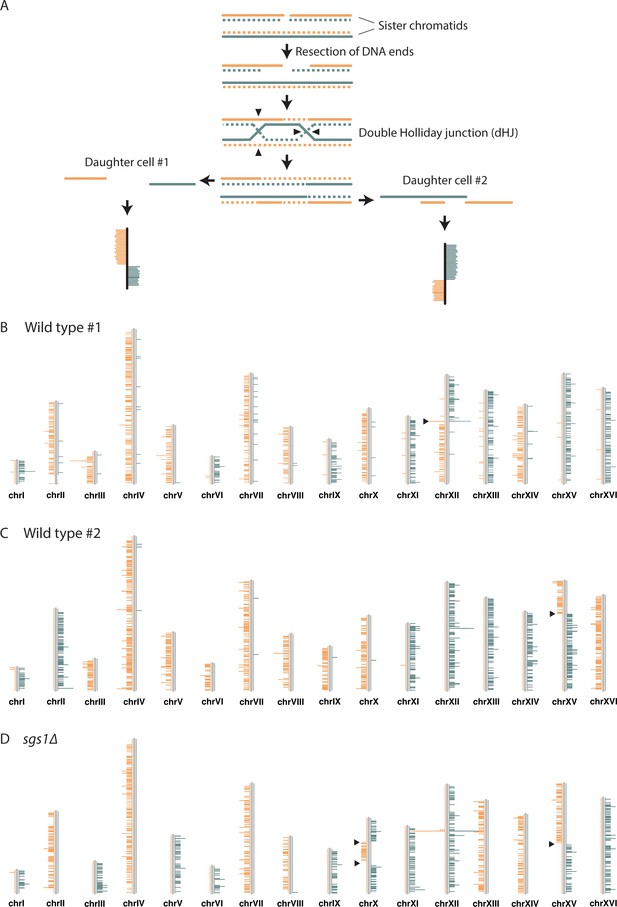
Detection of SCE events using Strand-seq.
(A) An SCE can occur as a result of DSB repair. Two sister chromatids, one of which has a DSB, are shown. The parental template DNA strands are depicted with solid lines, while the newly synthesized strands containing BrdU are depicted with dashed lines. The Watson and Crick strands are shown in orange and blue, respectively. DSB repair by SCR can lead to the formation of a double Holliday junction (dHJ). Resolution of the dHJ by structure-specific endonucleases will result in either a noncrossover (not shown) or a crossover. The resulting sister chromatids are then segregated to two different daughter cells. In the current Strand-seq protocol, only one daughter cell is isolated and analyzed. The BrdU-containing strands are nicked during library preparation, resulting in the sequencing of only parental strands. Sequence reads are mapped to either side of a chromosome ideogram. An SCE results in a switch from Watson to Crick reads along the chromosome. Note: the small gap between the parental strands in daughter cell #1 and the small overlap of the parental strands in daughter cell #2 are too small to be detected with Strand-seq. (B) An example of a wild-type Strand-seq library. Ideograms of the 16 yeast chromosomes are shown. Orange and blue lines correspond to reads aligning to the Watson and Crick strands, respectively. This cell inherited either the parental Watson strand or the parental Crick strand for each chromosome, except chromosome XII. A switch from Watson to Crick reads can be seen for chromosome XII (black arrowhead), indicating that an SCE event has occurred. (C) A second example of a wild-type Strand-seq library. An SCE event was detected on chromosome XV. (D) An example of an sgs1∆ Strand-seq library. Three SCE events were detected in this library: two on chromosome X and one on chromosome XV.
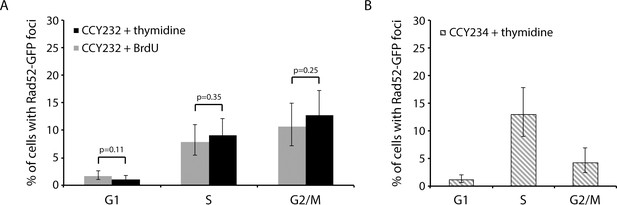
Incorporation of BrdU does not increase Rad52 focus formation during the first cell cycle.
(A) The percentages of G1, S, and G2/M cells with at least one Rad52-GFP focus are shown for CCY232 (isogenic to the E17 wild-type Strand-seq strain, except with RAD52-GFP) grown in the presence of thymidine or BrdU for 90 min (thymidine: G1 = 18/1058, S = 32/404.5, G2/M = 25.5/240.5; BrdU: G1 = 10.5/1009, S = 37.5/415, G2/M = 35/275). Numbers are averages of cells counted by two people independently. (B) As an additional control, the percentages of G1, S, and G2/M cells with at least one Rad52-GFP focus were determined for CCY234 (a wild-type ‘non-Strand-seq’ strain that expresses Rad52-GFP; G1 = 9.5/823, S = 31/240, G2/M = 13.5/318.5), and are similar to previously published data (Lisby et al., 2001). Error bars show 95% confidence intervals. P-values were calculated using Fisher’s exact test.
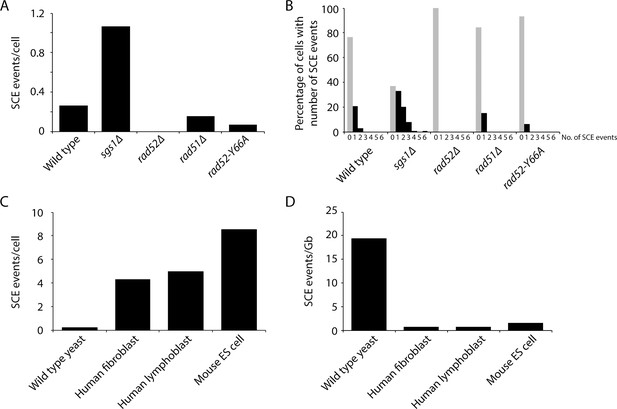
Measurement of spontaneous SCE genome-wide in single cells.
(A) Number of SCE events per cell for the indicated genotypes. (B) Percentage of cells with the indicated number of SCE events for each genotype. (C) Number of SCE events per cell for wild-type yeast, human fibroblasts, human lymphoblasts, and mouse ES cells. (D) SCE events per gigabase of DNA for wild-type yeast, human fibroblasts, human lymphoblasts, and mouse ES cells.
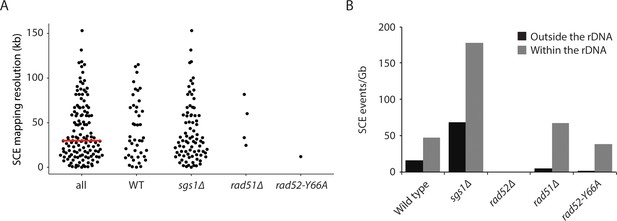
Mapping SCE events.
(A) Mapping resolution of SCE events in all yeast Strand-seq libraries and by genotype. The red line shows the median mapping resolution for all libraries. SCE events within the rDNA locus are excluded from this analysis because it is not possible to determine where within the rDNA an SCE event has occurred. (B) SCE events per gigabase of DNA, for either the entire genome excluding the rDNA locus or only considering the rDNA locus, were plotted for the indicated genotypes.
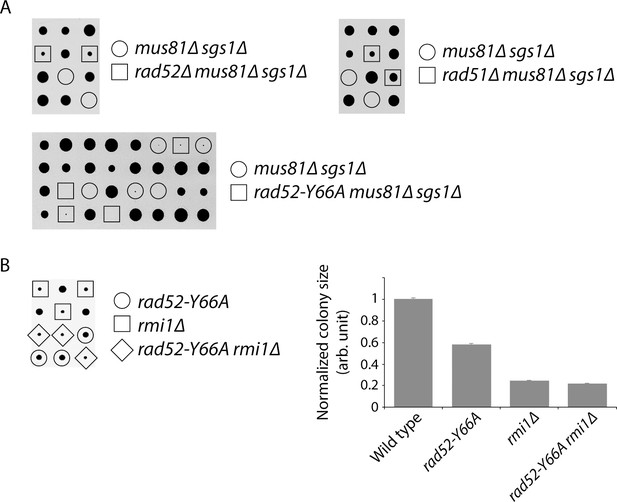
Abolishing the strand annealing activity of Rad52 does not suppress mus81∆ sgs1∆ synthetic lethality or rmi1∆ slow growth.
(A) Representative tetrads derived from the sporulation of MCY736, MCY737, and MCY773 are shown. (B) Representative tetrads derived from the sporulation of CCY198 are shown. Colony sizes for the indicated genotypes were measured and normalized to wild type. Mean ±SEM is shown. Lack of suppression of mus81∆ sgs1∆ synthetic lethality and rmi1∆ slow growth was also observed using another rad52 class C mutant: rad52-R70A (data not shown).
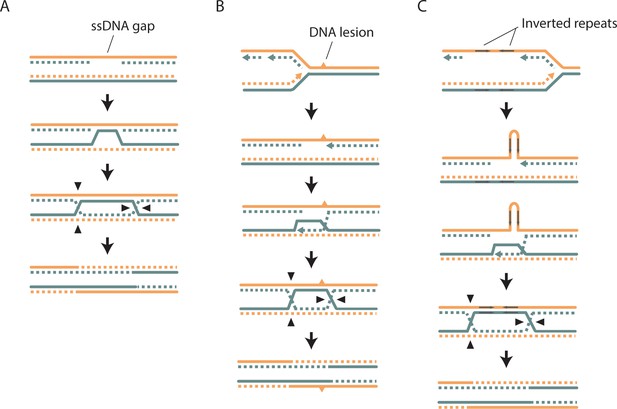
Models of SCE that do not involve a DSB.
As in Figure 1A, the parental template DNA strands are depicted with solid lines, while the newly synthesized strands are depicted with dashed lines. The Watson and Crick strands are shown in orange and blue, respectively. (A) An SCE could be generated from the repair of an ssDNA gap. This could proceed via inverse strand exchange, where Rad52 forms a complex with dsDNA and promotes strand exchange with a homologous ssDNA sequence independently of Rad51 (Mazina et al., 2017). (B) A DNA lesion on one of the parental template strands can cause template switching, where nascent DNA is used as a template for DNA replication, and could result in SCE. (C) As in B), but the ‘DNA lesion’ is an inverted DNA repeat forming a hairpin loop.
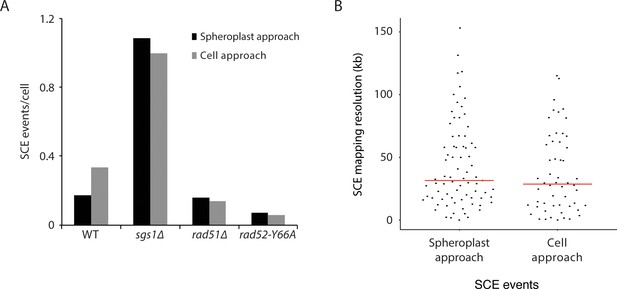
The spheroplast-sorting approach compared to the cell-sorting approach in terms of (A) number of SCE events per cell (divided by genotype) and (B) SCE mapping resolution.
https://doi.org/10.7554/eLife.30560.010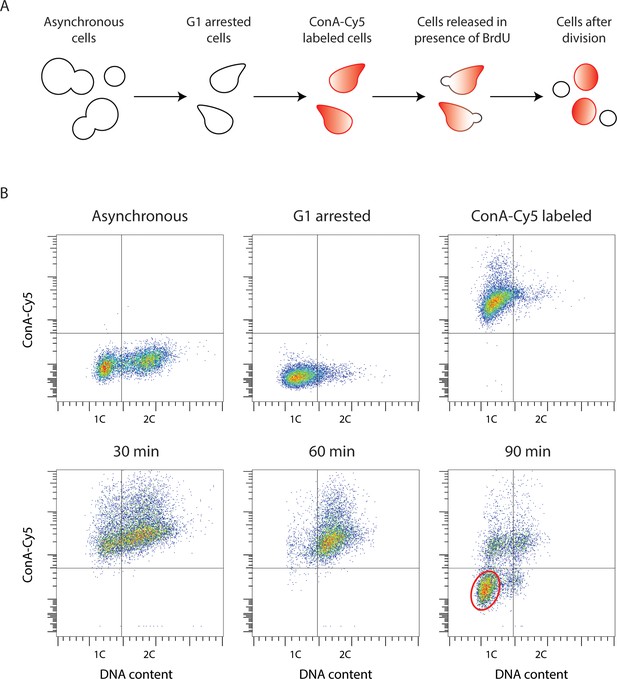
Yeast Strand-seq via the 'single cell sorting' approach.
(A) Scheme of the ‘single cell sorting’ approach for yeast Strand-seq. (B) Representative flow cytometry scatter plots corresponding to the scheme depicted in (A). Cells appearing in the lower left quadrant in the 90 min post-release sample are newly divided daughter cells to be sorted. A typical window for sorting is shown by the red oval.
Tables
Comparison of SCE by genotype.
https://doi.org/10.7554/eLife.30560.005| Genotype | No. of cells analyzed | No. of SCEs (SCEs/cell) | No. of SCEs at rDNA | SCEs/Gb (outside rDNA) | SCEs/Gb (within rDNA) |
|---|---|---|---|---|---|
| Wild type | 218 | 57 (0.26) | 14 | 16.3 | 47.0 |
| sgs1∆ | 103 | 110 (1.07) | 25 | 68.2 | 177.8 |
| rad52∆ | 27 | 0 (0) | 0 | 0 | 0 |
| rad51∆ | 65 | 10 (0.15) | 6 | 5.1 | 67.6 |
| rad52-Y66A | 76 | 5 (0.07) | 4 | 1.1 | 38.6 |
Yeast strains used in this study.
https://doi.org/10.7554/eLife.30560.009| Strain name | Relevant genotype | Source |
|---|---|---|
| E17 | MATa ADE2 cdc21::kanMX leu2::LEU2-GAL-hENT1 LYS2 RAD5 trp1::TRP1-GAL-dNK ura3-1 | Peter Thorpe |
| CCY232 | MATa ADE2 cdc21::kanMX leu2::LEU2-GAL-hENT1 LYS2 RAD5 trp1::TRP1-GAL-dNK ura3-1 RAD52-GFP::HIS3M × 6 | This study |
| CCY234 | MATa ADE2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 RAD52-GFP::HIS3M × 6 | This study |
| CCY118 | MATa ADE2 cdc21::natMX leu2::LEU2-GAL-hENT1 LYS2 RAD5 trp1::TRP1-GAL-dNK ura3-1 sgs1∆kanMX | This study |
| CCY193 | MATa ADE2 cdc21::natMX leu2::LEU2-GAL-hENT1 LYS2 RAD5 trp1::TRP1-GAL-dNK ura3-1 rad52∆kanMX | This study |
| CCY150 | MATa ADE2 cdc21::kanMX leu2::LEU2-GAL-hENT1 LYS2 RAD5 trp1::TRP1-GAL-dNK ura3-1 rad51∆natMX | This study |
| CCY182 | MATa ADE2 cdc21::kanMX leu2::LEU2-GAL-hENT1 LYS2 RAD5 trp1::TRP1-GAL-dNK ura3-1 rad52-Y66A | This study |
| MCY736 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 RAD5/RAD5 mus81ΔkanMX/MUS81 rad52∆/RAD52 sgs1ΔHIS3/SGS1 | This study |
| MCY737 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 RAD5/RAD5 mus81ΔkanMX/MUS81 rad51∆natMX/RAD51 sgs1ΔHIS3/SGS1 | This study |
| MCY773 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 RAD5/RAD5 rad52-Y66A::hphMX/RAD52 sgs1ΔHIS3/SGS1 | This study |
| CCY198 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 RAD5/RAD5 rad52-Y66A/RAD52 rmi1ΔkanMX/RMI1 | This study |
| MCY735 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 RAD5/RAD5 mus81ΔkanMX/MUS81 rad52-R70A/RAD52 sgs1ΔHIS3/SGS1 | This study |
| CCY196 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 RAD5/RAD5 rad52-R70A/RAD52 rmi1ΔkanMX/RMI1 | This study |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30560.012





