Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands
Figures
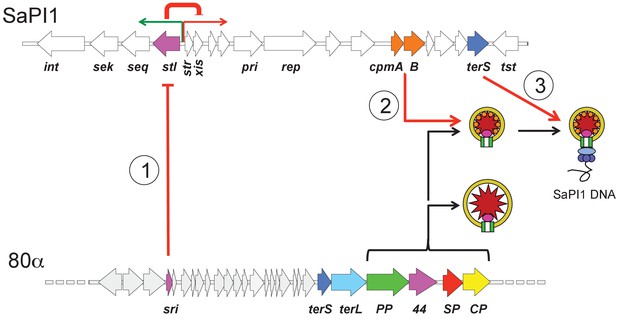
Schematic overview of SaPI1’s molecular piracy of 80α.
Only part of the 80α genome is shown for clarity, and relevant genes are labeled: sri, SaPI1 derepressor; terS, terminase small subunit; terL, terminase large subunit; PP, portal protein; 44, minor capsid protein gp44; SP, scaffolding protein; CP, major capsid protein. First, the 80α-encoded protein Sri interacts with the SaPI1 master repressor Stl (1), causing derepression of the rightwards operon that includes the transcriptional activator str, the excisionase xis and the replicase (pri and rep). Second, the SaPI1-encoded proteins CpmA and CpmB re-direct the capsid assembly pathway to form small capsids (2). Finally, the SaPI1-encoded TerS interacts with 80α-encoded TerL to cause specific packaging of SaPI1 DNA (3). Also labeled are the SaPI1 integrase (int), genes encoding staphylococcal enterotoxins K (sek) and Q (seq), and the toxic shock syndrome toxin (tst) gene.
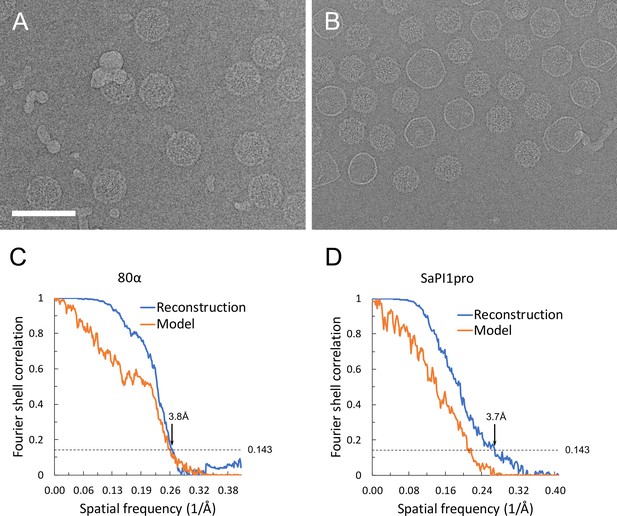
Cryo-EM data.
(A, B) Electron micrographs of 80α (A) and SaPI1 (B) procapsids. Scale bar, 100 nm. (C, D) Fourier Shell Correlation (FSC) curves between maps calculated from the two half datasets (blue) and between the model and the map (orange) for 80α (A) and SaPI1 (B). The resolution limits at 0.143 cutoff (dashed line) are indicated. The discrepancy between the model and the map for SaPI1 is likely due to the disordered and unmodeled portions of the CpmB protrusions.
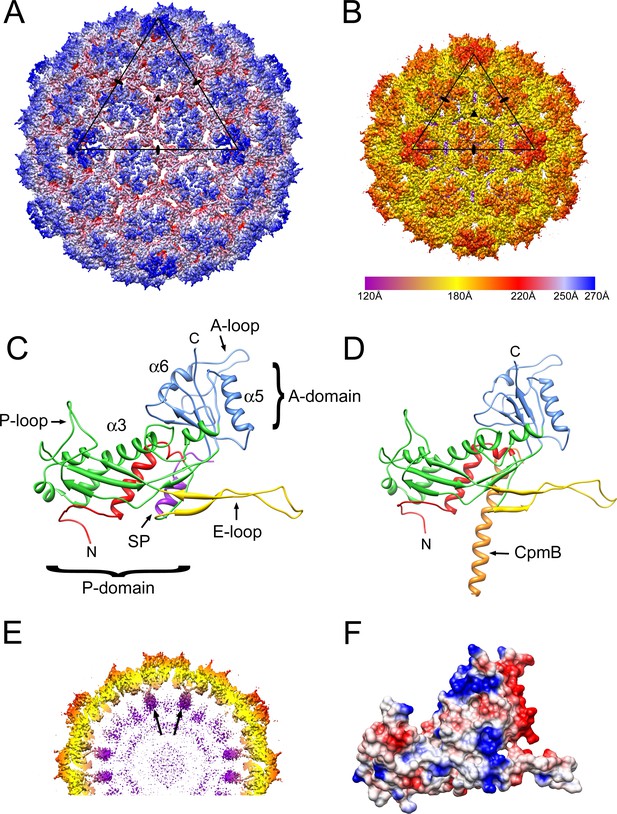
Reconstruction of 80α and SaPI1 procapsids.
(A, B) Isosurface representation of the 80α (A) and SaPI1 (B) reconstructions, colored radially on an absolute scale (color bar). The triangles represent one icosahedral face, delimited by three fivefold axes. Icosahedral twofold and threefold symmetry axes are indicated by ovals and triangles, respectively. (C, D) Ribbon representations of the C subunits of the 80α (C) and SaPI1 (D) atomic models. N-arm, red; P domain, green; E-loop, yellow; A-domain, blue. SP (in 80α), purple; CpmB (in SaPI1), orange. Locations of α-helices α3 (the spine helix), α5 and α6, and the A-loop are also indicated in (C). (D) Isosurface representation of a 30 Å thick slice through the SaPI1 procapsid map, showing the protruding domains on the inside of the shell (arrows), colored radially on the same scale as in (B). (G) Electrostatic surface of the 80α CP subunit C, colored according to charge (blue, most positive; red, most negative).
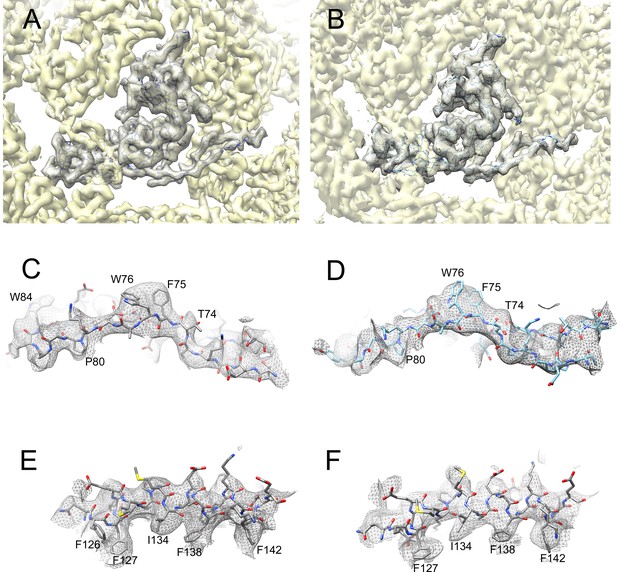
Electron density and model.
(A, B) Isosurface representation (mesh) of one whole subunit from 80α (A) and SaPI1 (B), with the corresponding CP model in backbone representation. The surrounding density is shown as a semi-transparent yellow isosurface. (C–F) Details of density and atomic model from 80α (C, E) and SaPI1 (D, F), corresponding to residues 68–84 in the E-loop (C, D) and 125–144 in the spine helix (α3) (E, F). Pertinent residues are labeled.
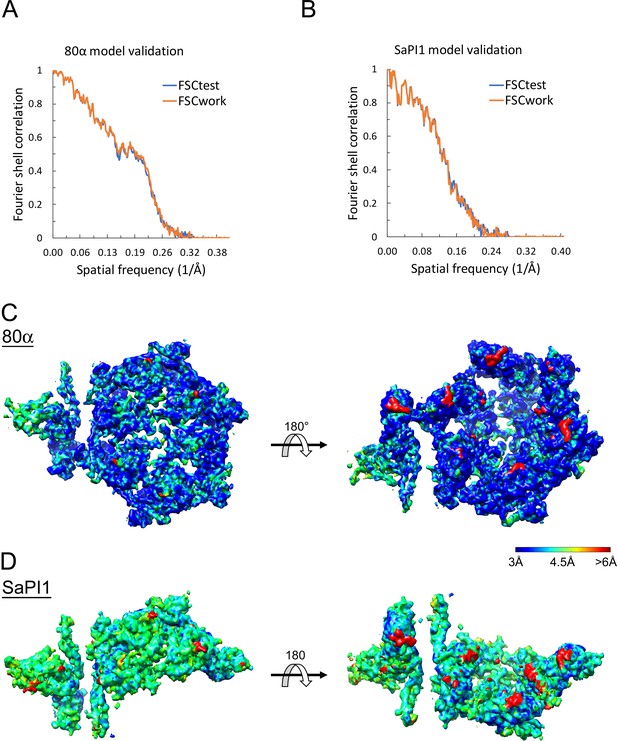
Data analysis.
(A, B) FSC curves showing the correspondence of the model against the test dataset (FSCtest, blue) and the working dataset (FSCwork, orange), after refinement against only the working data set for the 80α (A) and SaPI1 (B) procapsid reconstructions. (C, D) Isosurface representation of one asymmetric unit from the 80α (C) and SaPI1 (D) procapsid maps, colored by local resolution from 3 Å to >6 Å according to the color bar, calculated using ResMap (Kucukelbir et al., 2014).
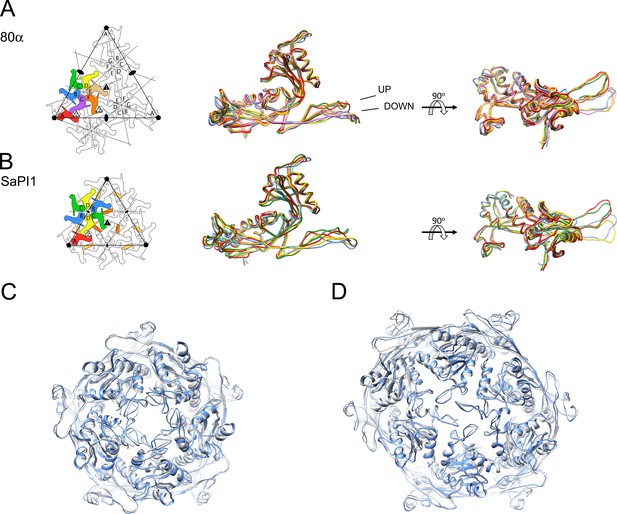
Comparison of CP subunits.
(A, B) Superposition of the backbones of the seven subunits of 80α (A) and the four subunits of SaPI1 (B). Panels on the right are rotated by 90°. The two distinct conformations of the E-loop (‘UP’ and ‘DOWN’) are indicated on the 80α superposition. The subunits are colored according to the schematic diagrams on the left (A, red; B, blue; C, green; D, yellow; E, tan; F, orange; G, purple). The orange ovals in (B) represent the CpmB dimers. (C, D) Superpositions of the pentamers (C) and hexamers (D) of 80α (gray) and SaPI1 (blue), in ribbon representation.
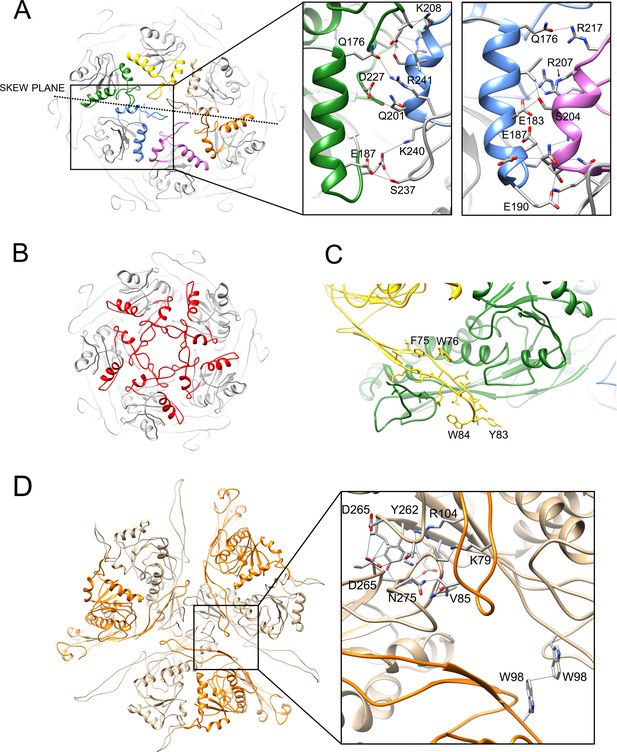
Interactions between capsid proteins.
(A, B) Ribbon representation of the 80α pentamer (A) and hexamer (B), showing the detailed interactions between the A-domains. Helices α5 and α6 and the A-loop are colored according to the color scheme in Figure 4A. The skew plane between B-C and E-F subunits is indicated by the dashed line. Pertinent residues are shown in stick representation and labeled in the expanded view on the right, showing the two distinct types of A-domain interactions. (C) Detail of the 80α model, showing the interactions between the E-loop of subunit D (yellow) and the P-domain and P-loop of subunit C (green). (D) Detail of the trimer interaction at the icosahedral threefold axis showing three E subunits (tan) and the corresponding adjacent F subunits (orange). Key residues involved in the threefold interaction, as well as W98, are indicated in stick model in the expanded view.
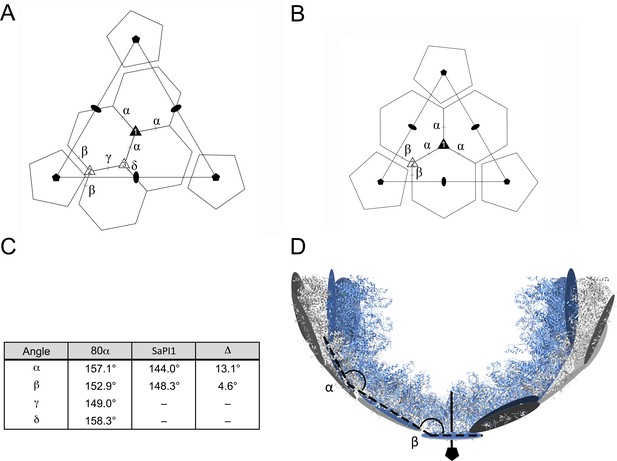
Comparison of capsomer angles in 80α and SaPI1.
(A, B) Schematic diagrams showing the organization of hexamers and pentamers in the T = 7 (A) and T = 4 (B) lattices. The dihedral angles between hexamers are defined by the Greek letters α–δ. Type 1, 2 and 3 threefold axes are indicated. (C) Table of the calculated interior inter-capsomeric dihedral angles in 80α and SaPI1. The Δ column indicates the difference between corresponding angles. (D) Ribbon representation of a slab through one half of the 80α (gray) and SaPI1 (blue) shells, showing how the difference in inter-capsomer angles are propagated through the lattice. The models were aligned at one fivefold vertex (shown at bottom of diagram) and the planes representing the capsomers (gray and blue circles) and the resulting α and β angles are shown.
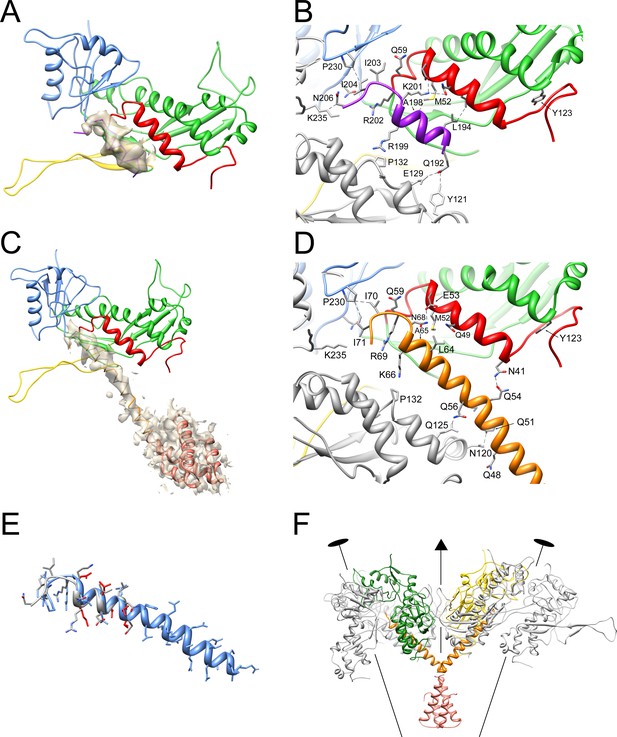
Interaction of CP with SP and CpmB.
(A) Ribbon diagram of CP and SP from subunit C of the 80α procapsid with the density corresponding to SP superimposed, colored as in Figure 3E. (B) Detail of the SP-CP interaction, with relevant residues highlighted (stick representation) and labeled. Key interactions (<4 Å) are indicated by stippled lines. (C) Ribbon diagram of CP and CpmB from subunit C in the SaPI1 procapsid showing the density corresponding to CpmB, colored as in Figure 3F. The NMR structure of the N-terminal dimerization domain of CpmB (salmon) has been placed in the density, but could not be modeled accurately. (D) Detail of CpmB-CP interaction with relevant residues and distances (<4 Å) indicated. (E) Superposition of CpmB (blue) and SP (gray). Side chains in CpmB for residues that differ from their equivalents in SP are shown in red. (F) Ribbon diagram showing the interaction of one CpmB dimer (orange and salmon) with two adjacent hexameric capsomers. The C and D subunits are colored (green and yellow, respectively); other subunits are in gray. The positions of two twofold and one threefold symmetry axis are indicated.
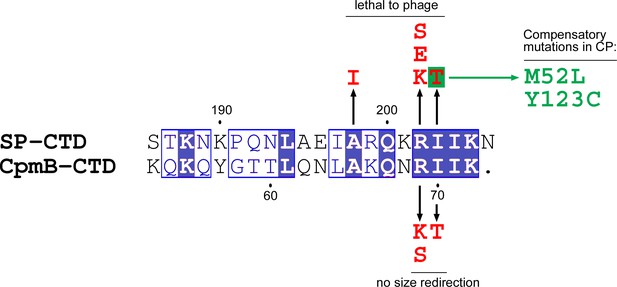
Sequence alignment of the C-terminal domains of SP and CpmB.
Size redirection negative mutations in CpmB and lethal mutations in SP are indicated (red). The mutations in CP that suppressed the I203T mutant are listed (green). Alignment generated in ClustalW and rendered with ESPript 2.0.
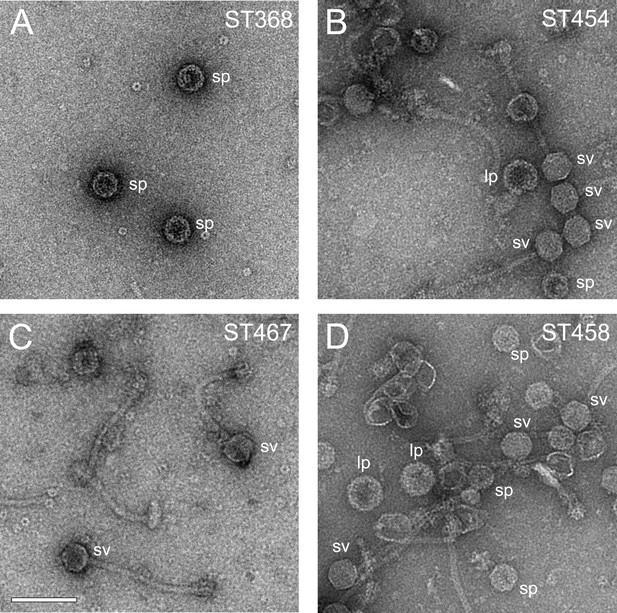
Electron micrographs of negatively stained particles from mutant lysates.
(A) Strain ST368 (80α SP I203T + wildtype SaPI1); (B) strain ST454 (80α SP A198I + wildtype SaPI1; (C) strain ST467 (80α SP-CpmBCTD + wildtype SaPI1); (D) strain ST458 (80α wildtype + SaPI1 CpmB-SPCTD). Examples of small procapsids (sp), small virions (sv), and large procapsids (lp) are indicated on the images. Scale bar denotes 100 nm.
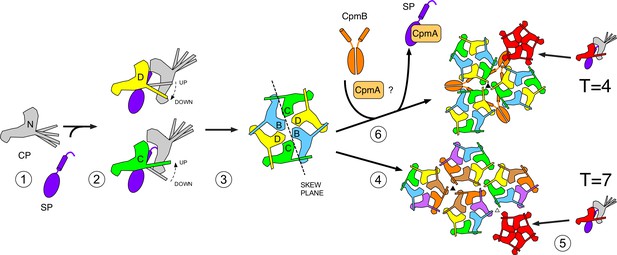
Schematic model for scaffolding competition-mediated capsid size redirection.
In this scheme, SP is needed for the correct folding of CP (1), which is only stable as an oligomer. The E-loop conformation is ‘up’ or ‘down’ depending on its binding to a neighboring CP subunit (2). SP might be involved in this conformational switching. Subunits are assembled into hexamers (and possibly other oligomers), which are independent of the presence or absence of CpmB (3). The curvature imposed on the nascent shell by the trivalent interaction of hexamers (4) forces the incorporation of pentamers into the lattice at specific points (5), leading to a T = 7 shell. The presence of CpmB dimers imposes a sharper angle between hexamers (6), leading to the more frequent incorporation of pentamers and the formation of a the smaller, T = 4 shell. CpmA is thought to facilitate the removal of SP to provide access for CpmB (6).
Tables
Root-mean square deviation (RMSD) values (in Å) between subunit pairs within 80α, within SaPI1 and between 80α and SaPI1.
Values for ‘unpruned’ structures include all atoms when determining the best fit. Values for ‘pruned’ structures only include the best matching residue pairs. Pruning was done in UCSF Chimera using standard parameters. For 80α, pairs of subunits related across the quasi-twofold axis are highlighted in green. Comparison of subunits for which the E-loops are in the ‘UP’ conformation are highlighted in blue.
| Root mean square deviation between CP subunits | |||||
|---|---|---|---|---|---|
| Pruned | Unpruned | ||||
| # res. | RMSD (Å) | # res. | RMSD (Å) | ||
| 80α procapsid | |||||
| subA | subB | 215 | 0.986 | 284 | 2.927 |
| subA | subC | 250 | 0.897 | 284 | 1.521 |
| subA | subD | 239 | 0.943 | 284 | 2.902 |
| subA | subE | 218 | 0.989 | 284 | 2.900 |
| subA | subF | 252 | 0.899 | 284 | 1.533 |
| subA | subG | 239 | 1.024 | 284 | 2.878 |
| subB | subC | 224 | 0.881 | 284 | 3.072 |
| subB | subD | 229 | 0.955 | 284 | 1.901 |
| subB | subE | 251 | 0.727 | 284 | 1.506 |
| subB | subF | 219 | 0.907 | 284 | 3.244 |
| subB | subG | 232 | 0.909 | 284 | 1.686 |
| subC | subD | 241 | 0.853 | 284 | 3.002 |
| subC | subE | 225 | 0.923 | 284 | 3.108 |
| subC | subF | 271 | 0.764 | 284 | 0.987 |
| subC | subG | 237 | 0.860 | 284 | 2.974 |
| subD | subE | 231 | 1.001 | 284 | 1.715 |
| subD | subF | 246 | 0.781 | 284 | 3.195 |
| subD | subG | 262 | 0.704 | 284 | 1.302 |
| subE | subF | 220 | 0.979 | 284 | 3.257 |
| subE | subG | 236 | 0.981 | 284 | 1.654 |
| subF | subG | 242 | 0.847 | 284 | 3.104 |
| SaPI1 procapsid | |||||
| subA | subB | 218 | 1.047 | 284 | 2.656 |
| subA | subC | 247 | 1.092 | 284 | 1.572 |
| subA | subD | 223 | 1.108 | 284 | 2.862 |
| subB | subC | 220 | 1.040 | 284 | 2.865 |
| subB | subD | 231 | 1.040 | 284 | 1.633 |
| subC | subD | 242 | 0.921 | 284 | 2.669 |
| SaPI1 vs. 80α | |||||
| SaPI1 | 80α | ||||
| subA | subA | 259 | 0.920 | 284 | 1.495 |
| subB | subB | 250 | 0.926 | 284 | 1.443 |
| subB | subE | 257 | 0.823 | 284 | 1.380 |
| subC | subC | 268 | 0.935 | 284 | 1.165 |
| subC | subF | 268 | 0.997 | 284 | 1.190 |
| subD | subD | 247 | 0.948 | 284 | 1.536 |
| subD | subG | 262 | 0.939 | 284 | 1.170 |
List of contacts between SP and CP in the 80α procapsid (panel A, left) and between CpmB and CP in the SaPI1 procapsid (panel B, right).
Most contacts are within the same subunit (chain C), but some contacts are made with the adjacent subunit (chain B). Corresponding residues in SP and CpmB are on the same row. The assigned SP sequence starts on P191, while CpmB starts on Q48. A contact is defined as an interatomic distance of ≤4 Å. The table lists the specific residues that are involved in these contacts and enumerates the residues (# Res) and atoms (# Atoms) that contribute to these contacts. Residues that make the most contacts (≥5 interatomic pairs) are highlighted in green, while those that display ≤1 contacts are highlighted in red. ‘Intermediate’ contacts (2–4) are shown in yellow.
| A. 80α procapsid. contacts (≤4 Å) between SP (subunit C) and CP | B. SaPI1 procapsid. contacts (≤4 Å) between CpmB (subunit C) and CP | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SP | CP chain C | CP chain B | Res | Atoms | CpmB | CP chain C | CP chain B | Res | Atoms | ||
| Q | 48 | N120 | 1 | 5 | |||||||
| E | 49 | 0 | 0 | ||||||||
| E | 50 | 0 | 0 | ||||||||
| Q | 51 | E117, N120 | 2 | 13 | |||||||
| S | 52 | 0 | 0 | ||||||||
| K | 53 | 0 | 0 | ||||||||
| Q | 54 | N41 | 1 | 2 | |||||||
| K | 55 | E67 | E117, Y121 | 3 | 7 | ||||||
| Q | 56 | Q125 | 1 | 11 | |||||||
| Y | 57 | 0 | 0 | ||||||||
| P | 191 | 0 | 0 | G | 58 | 0 | 0 | ||||
| Q | 192 | E129, Y121 | 2 | 5 | T | 59 | 0 | 0 | |||
| N | 193 | 0 | 0 | T | 60 | 0 | 0 | ||||
| L | 194 | T45, L48, I253 | 3 | 6 | L | 61 | T45 | 1 | 1 | ||
| A | 195 | Y63 | 1 | 1 | Q | 62 | Y63, E64, P65 | 3 | 17 | ||
| E | 196 | 0 | 0 | N | 63 | 0 | 0 | ||||
| I | 197 | T45, Q49, M52 | 3 | 4 | L | 64 | Q49 | 1 | 7 | ||
| A | 198 | M52, Y63 | 2 | 4 | A | 65 | M52, Y63 | 2 | 5 | ||
| R | 199 | E50, P32 | 2 | 5 | K | 66 | P132 | 1 | 2 | ||
| Q | 200 | 0 | 0 | Q | 67 | 0 | 0 | ||||
| K | 201 | Q49, E53 | 2 | 6 | N | 68 | M52, E53 | 2 | 5 | ||
| R | 202 | Q59, L60, G61, K62, G247 | 5 | 4 | R | 69 | M58, Q59, L60, G61 | 4 | 8 | ||
| I | 203 | Q59, V232, P230 | 3 | 7 | I | 70 | Q59, P230 | 2 | 6 | ||
| I | 204 | L60, N194, A195, P230 | 4 | 11 | I | 71 | N194, P230 | K235 | 3 | 3 | |
| K | 205 | 0 | 0 | K | 72 | K235 | 1 | 1 | |||
| N | 206 | Y139, K235 | 2 | 6 | – | – | |||||
Genetic analysis of 80α and SaPI1, listing the S. aureus strains and the genotypes of the 80α prophages and SaPI1 elements that they harbor.
SaPI1 also includes the tst::tetM insertion that confers tetracycline resistance. The corresponding phage titers (PFU/ml) and tetM transducing titers (TU/ml) are from filtered lysates resulting from induction with mitomycin C. Titers of mutations considered lethal or greatly impaired are shown in red; titers that are essentially wildtype are shown in green. WT = wildtype; N/D = not determined.
| Strain | Phage | SaPI1 | Phage titer | SaPI1 titer |
|---|---|---|---|---|
| RN10616 | WT | – | 8.70E + 10 | – |
| RN10628 | WT | WT | 5.10E + 08 | 1.33E + 08 |
| ST196 | SP R202K | – | <10 | – |
| ST197 | SP R202K | WT | N/D | <10 |
| ST278 | SP R202S | – | <10 | – |
| ST366 | SP R202S | WT | <10 | 1.67E + 01 |
| ST279 | SP R202E | – | <10 | – |
| ST367 | SP R202E | WT | N/D | <10 |
| ST358 | SP I203T | – | 1.00E + 02 | – |
| ST368 | SP I203T | WT | 1.71E + 02 | 6.67E + 06 |
| ST469 | SP I204L | – | 5.33E + 10 | – |
| ST417 | SP A198I | – | 1.00E + 01 | – |
| ST454 | SP A198I | WT | <10 | 4.18E + 06 |
| ST384 | CP M52L | – | 1.30E + 10 | – |
| ST415 | CP M52Q | – | 1.00E + 01 | – |
| ST481 | CP M52Q | WT | <10 | 3.00E + 01 |
| ST385 | CP Y123C | – | 1.47E + 10 | – |
| ST466 | SP::CpmBCTD | – | 2.70E + 09 | – |
| ST467 | SP::CpmBCTD | WT | 9.00E + 06 | 9.25E + 05 |
| ST468 | SP::CpmBCTD | CpmB::SPCTD | 6.80E + 06 | 9.00E + 05 |
| ST458 | WT | CpmB::SPCTD | 1.80E + 08 | 9.60E + 08 |
| ST465 | SP R202E | CpmB::SPCTD | <10 | 1.40E + 02 |
Additional files
-
Supplementary file 1
List of primers used to construct the plasmids in this study.
Restriction sites are underlined.
- https://doi.org/10.7554/eLife.30822.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30822.019





