TRAIN (Transcription of Repeats Activates INterferon) in response to chromatin destabilization induced by small molecules in mammalian cells
Figures
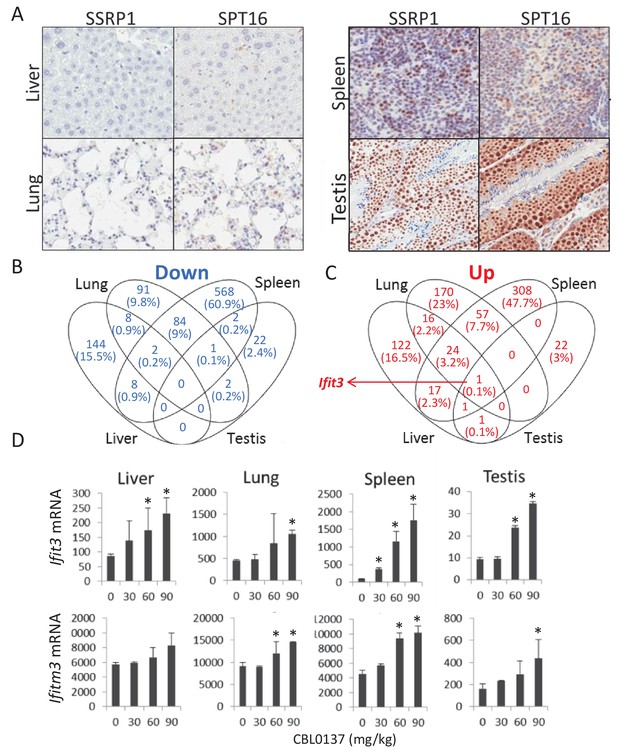
Effect of CBL0137 on transcription of genes in FACT-positive and -negative mouse tissues.
(A) IHC staining with SSRP1 antibody of liver and lung tissues with no detectable SSRP1 and spleen and testis expressing SSRP1. (B, C) Analyses of changes in genes expression in different tissues of mice treated with 30, 60 or 90 mg/kg of CBL0137 using microarray hybridization. Venn diagrams of the lists of genes for which expression was downregulated (B) or upregulated (C) in response to CBL0137 in different organs in response to any of doses of CBL0137. (D) Dose-dependent upregulation of expression of Ifit3 and Ifitm3 genes in different organs. Mean normalized value of microarray hybridization signals of two biological replicates ± SD. Asterisks indicate conditions when expression was increased >1.5 folds with p-value<0.05.
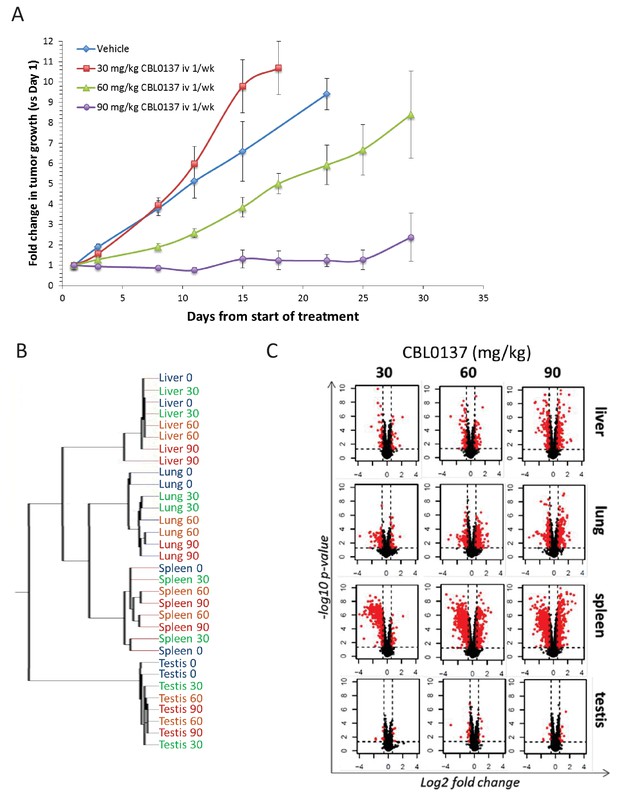
Effects of different doses of CBL0137 in tumor growth and gene expression in mice.
(A) Change in a volume of subcutaneous HepG2 tumors in SCID mice treated once a week with IV with vehicle (5% dextrose) or 30, 60 and 90 mg/kg of CBL0137 for 4 weeks. (B) Dendrogram of gene expression in different organs of mice treated with different doses of CBL0137 IV or control vehicle 24 hr before organ collection obtained using unsupervised hierarchical clustering. (C) Volcano plots of changes in gene expression in different organs of mice treated as in B.
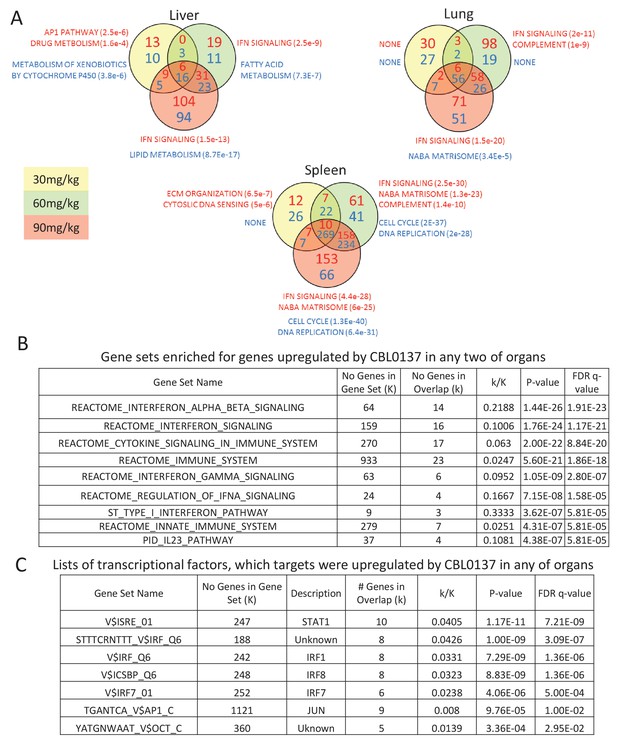
Activation of IFN response is detected in response to CBL0137 in different tissues of mice via GSEA.
(A) Venn diagrams showing the number of up (red font) or down (blue font) regulated genes in response to CBL0137 together with gene set names enriched for these genes. The p-value of overlap is shown in parentheses. (B) GSEA of curated signaling pathways using the list of genes upregulated in two or more tissues in response to any dose of CBL0137. (C) GSEA of targets of known transcriptional factors using the list of genes upregulated in any of tissues in response to any of doses of CBL0137.
-
Figure 2—source data 1
GSEA analysis of genes downregulated in response to treatment with 30 mg/kg (sheet 1), 60 mg/kg (sheet 2) and 90 mg/kg (sheet 3) of CBL0137 in liver.
- https://doi.org/10.7554/eLife.30842.006
-
Figure 2—source data 2
GSEA analysis of genes upregulated in response to treatment with 30 mg/kg (sheet 1), 60 mg/kg (sheet 2) and 90 mg/kg (sheet 3) of CBL0137 in liver.
- https://doi.org/10.7554/eLife.30842.007
-
Figure 2—source data 3
GSEA analysis of genes downregulated in response to treatment with 30 mg/kg (sheet 1), 60 mg/kg (sheet 2) and 90 mg/kg (sheet 3) of CBL0137 in lung.
- https://doi.org/10.7554/eLife.30842.008
-
Figure 2—source data 4
GSEA analysis of genes upregulated in response to treatment with 30 mg/kg (sheet 1), 60 mg/kg (sheet 2) and 90 mg/kg (sheet 3) of CBL0137 in lung.
- https://doi.org/10.7554/eLife.30842.009
-
Figure 2—source data 5
GSEA analysis of genes downregulated in response to treatment with 30 mg/kg (sheet 1), 60 mg/kg (sheet 2) and 90 mg/kg (sheet 3) of CBL0137 in spleen.
- https://doi.org/10.7554/eLife.30842.010
-
Figure 2—source data 6
GSEA analysis of genes upregulated in response to treatment with 30 mg/kg (sheet 1), 60 mg/kg (sheet 2) and 90 mg/kg (sheet 3) of CBL0137 in spleen.
- https://doi.org/10.7554/eLife.30842.011
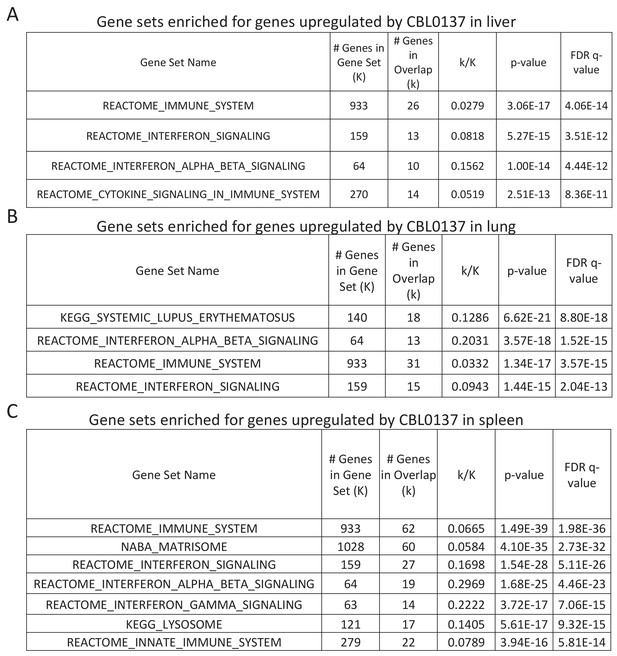
GSEA analysis of genes upregulated in liver (A), lung (B) and spleen (C) of mice treated with CBL0137.
https://doi.org/10.7554/eLife.30842.005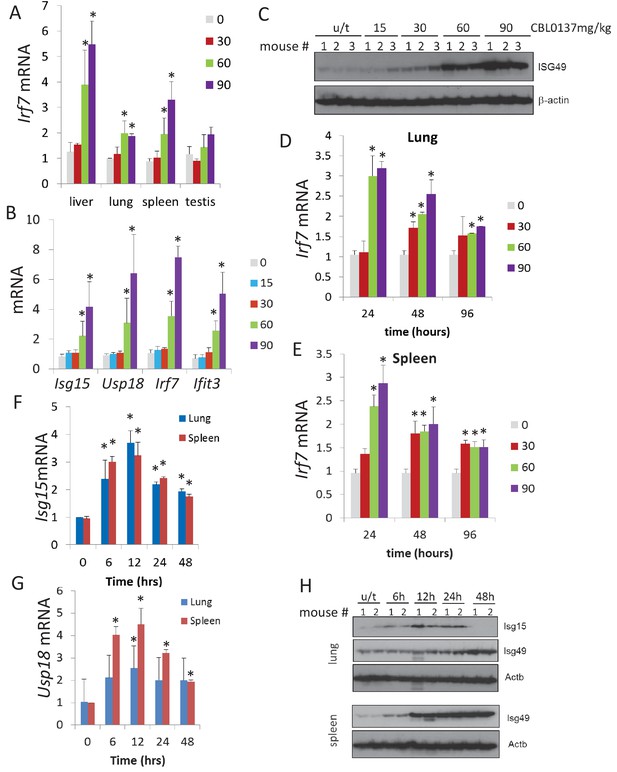
CBL0137 causes increased expression of IFN-responsive genes in different tissues of mice.
Quantitation of RT-PCR data (A, B, D, E, F, G) shown as fold change upon treatment with different doses of CBL0137 (mg/kg) comparing to vehicle-treated control. Mean values from three mice ± SD. Immunoblotting of mouse plasma (C) or tissue lysates (H). (A) Treatment of C57Bl/6 mice for 24 hr. B and C. Treatment of NIH Swiss mice for 24 hr. D - H. Different time treatment of C57Bl/6 mice. C and H – numbers indicate individual mice in each group. Bars – mean of two or three replicates +SD, asterisk – p<0.05 vs untreated control.
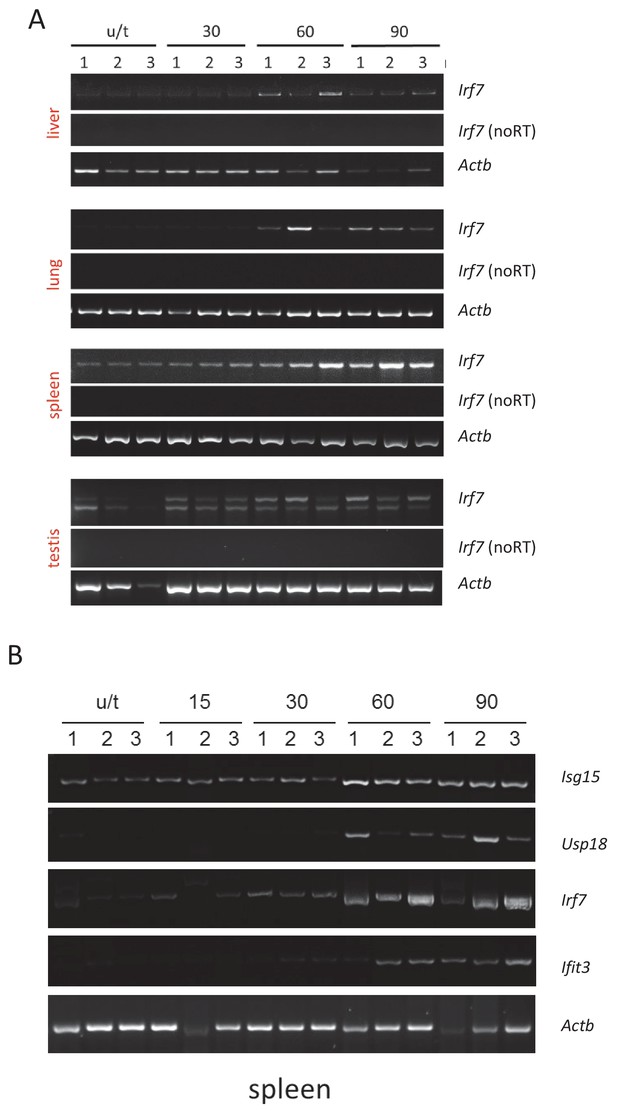
Images of RT-PCR reactions used for quantitation on Figure 3.
https://doi.org/10.7554/eLife.30842.013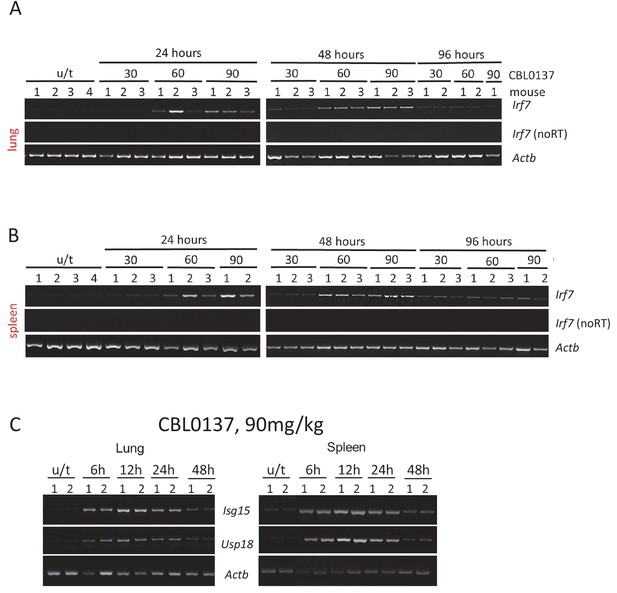
Images of RT-PCR reactions used for quantitation for Figure 3.
https://doi.org/10.7554/eLife.30842.014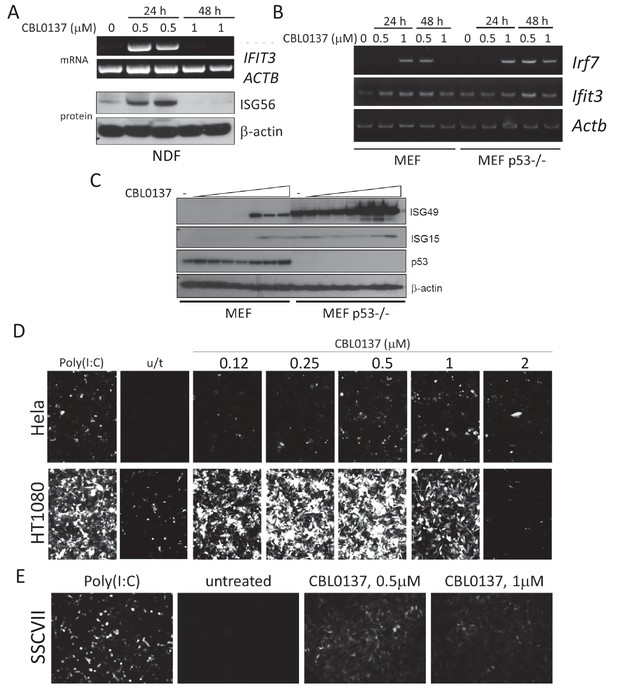
Induction of IFN response in human and mouse cells treated with CBL0137.
(A) RT-PCR and immunoblotting of mRNA and lysates from human normal diploid fibroblasts (NDF). ISG56 is a product of the human IFIT3 gene. (B) RT-PCR of mRNA from MEF, wild type or Trp53 (p53) deficient. (C) Immunoblotting of lysate of MEF treated with different doses of CBL0137 (0.03–2 μM) for 24 hr. D and E. Microphotographs of human (D) or mouse (E) tumor cells expressing ISRE-mCherry reporter, treated with different doses of CBL0137 for 48 hr. Poly(I:C) - polyinosinic:polycytidylic acid was used as a positive control for reporter activation at 50 μg/ml.
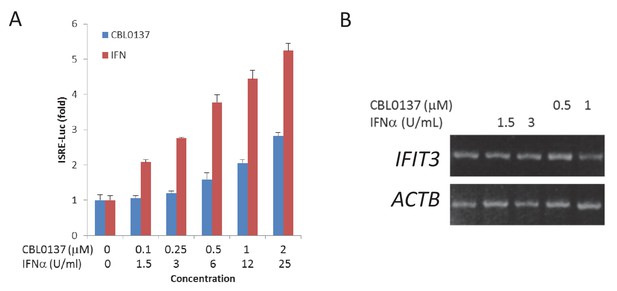
Comparison of the sensitivity of ISRE reporter assay with detection of the transcript of endogenous ISG, IFIT3 in human tumor cell line HCT116 transduced with lentiviral ISRE-Luc reporter.
Cells were treated for 36 hr with indicated doses of IFNα or CBL0137 and then either used for reporter assay (A) of for RNA isolation and RT-PCR (B).
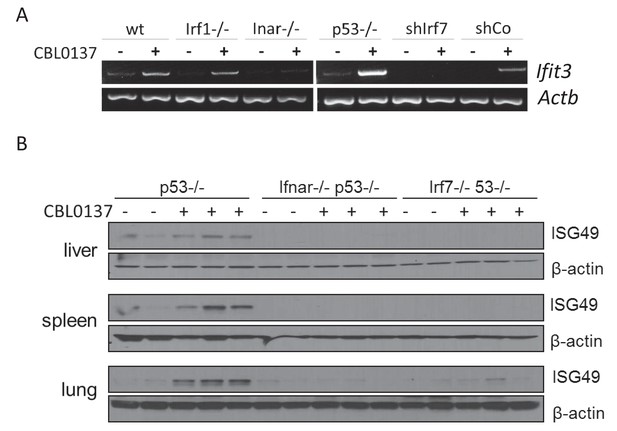
Induction of IFN-responsive gene by CBL0137 depends on IFN signaling.
(A) RT-PCR of RNA from MEF cells of different genotype or wild type MEF transduced with control shRNA or shRNA to Irf7 and treated for 24 hr with 0.5 μM of CBL0137. (B) Immunoblotting of tissue lysates from mice of different genotypes treated with vehicle control (n = 2 for each genotype) or 60 mg/kg CBL0137 (n = 3) 24 hr before tissue collection.
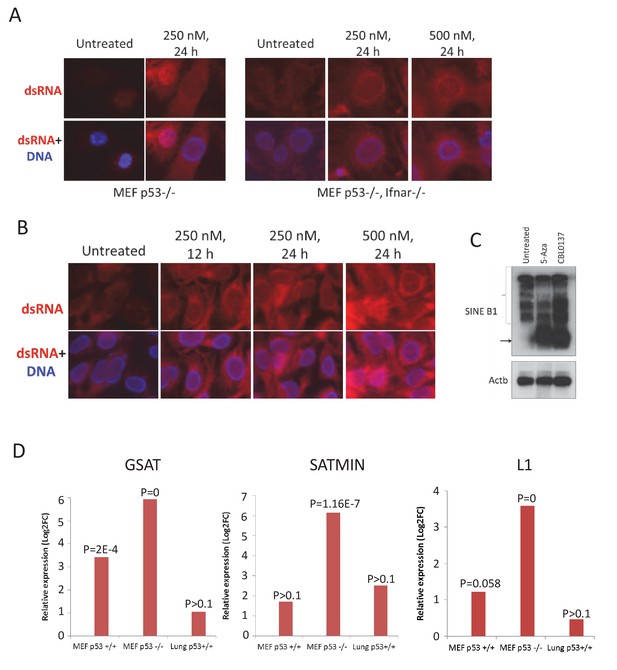
Elevated presence of double-stranded RNA in cells and tissues of mice treated with CBL0137.
Immunofluorescent staining of MEF (A) and human HT1080 (B) cells with antibody to dsRNA (red) and Hoeschst (blue). (C) Northern blotting of RNA from MEF (Trp53-/-) treated with either 5-Aza for 72 hr or CBL0137 for 48 hr and hybridized with B1 or Actb probes. (D) Increase in transcripts corresponding to major (GSAT) or minor (SATMIN) satellites or LINE1 in MEF cells or lung tissue of mice treated with either 1 μM or 60 mg/kg of CBL0137 for 24 hr. Bars are relative increase of the transcripts abundance in treated versus untreated samples. p=FDR corrected p-value (see Materials and methods for details).
-
Figure 6—source data 1
Analysis of expression of repetitive elements in control and CBL0137 treated wild type of Trp53-/- MEF cells of lungs of wild-type mice via RNA-seq.
- https://doi.org/10.7554/eLife.30842.020
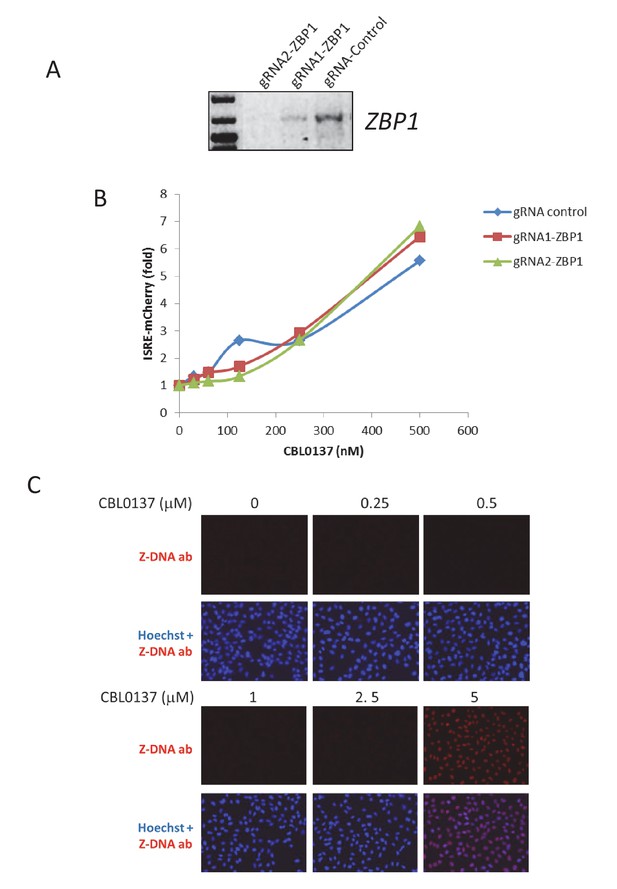
INF induction by CBL0137 does not depend on ZBP1.
(A) RT-PCR with primers to ZBP1 of mRNA from Hela cells in which ZBP1 gene was inactivated with CRISP-Cas9 recombinase. Two different gRNA to ZBP1 and one control were used. (B) Fluorescent intensity of ISRE-mCherry reporter in Hela cells control or with inactivated ZBP1 treated with CBL0137 for 48 hr. (C) Immunofluorescent staining of Hela cells treated with CBL0137 for 1 hr with antibody to Z-DNA (red) or Hoechst (blue).
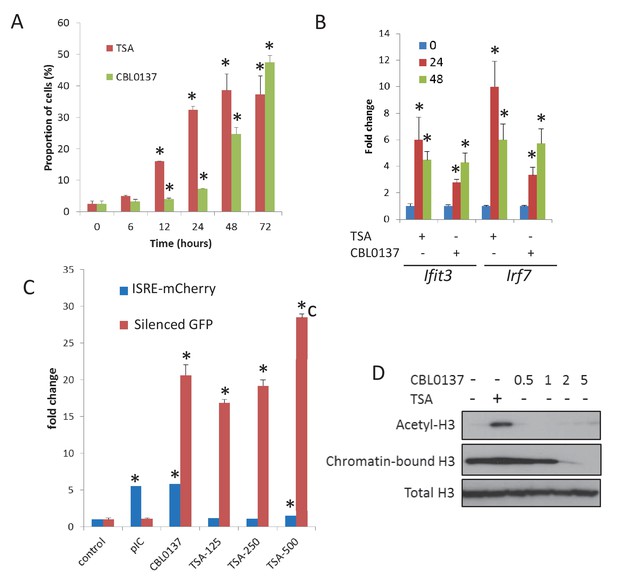
Chromatin opening is accompanied by TRAIN.
(A) Increase in the proportion of cells expressing GFP from silenced viral promoter in Hela cells treated with either TSA (250 nM) or CBL0137 (500 nM). (B) Changes in the abundance of Ifit3 and Irf7 mRNAs in MEF treated with 500 nm of TSA or CBL0137 for 24 or 48 hr. Quantitation of RT-PCR, mean of two experiments ± SD. All changes are significant versus untreated control (p<0.05). (C) Change in the proportion of Hela-TI-ISRE-mCherry cells positive for mCherry and GFP after 48 hr treatment with 10 μg/ml of polyI-C (pIC), 500 nM of CBL0137 or 125–500 nM of TSA. (D) Immunoblotting of extracts of Hela cells treated with TSA (200 nM) or different doses of CBL0137 (μM) for 24 hr. Acetyl H3 and total H3 were detected in total nuclear extracts, chromatin bound H3 - in chromatin pellet, washed from nucleoplasm with 450 mM NaCl buffer. Bars are mean of two replicates ± SD. Asterisk - p<0.05 vs untreated control.
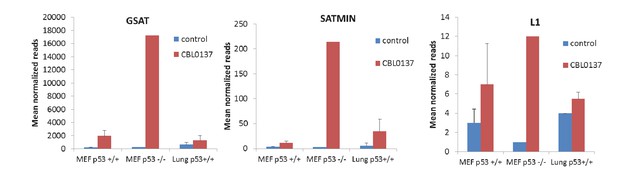
Abundance of transcripts corresponding to major (GSAT) or minor (SATMIN) satellites or LINE1 per millions of reads of RNA-seq of total RNA from MEF cells from Trp53 wild type (p53+/+) or Trp53-null (p53-/-) mice or lung tissue of Trp53 wild-type mice treated with either 1 μM or 60 mg/kg of CBL0137 for 24 hr.
https://doi.org/10.7554/eLife.30842.022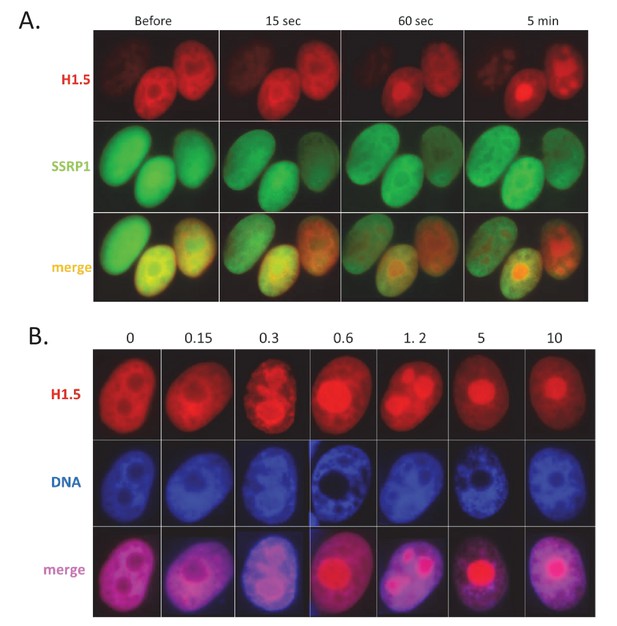
CBL0137 treatment leads to displacement of histone H1.
Live cell imaging of HT1080 cells expressing mCherry-tagged H1.5 and GFP-tagged SSRP1. (A) Images of nuclei of the same cells at different moments after addition of 1 μM CBL0137 to cell culture medium. (B) Images of nuclei of cells treated with different concentrations of CBL0137 (μM) for 1 hr.
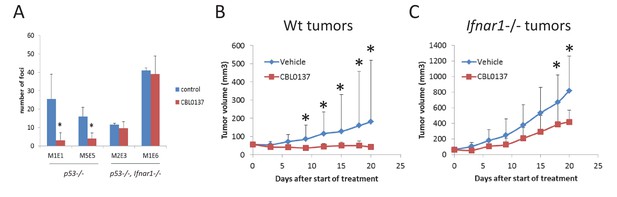
Input of IFN response into anti-cancer activity of CBL0137.
(A) Treatment of cells transduced with H-RasV12 with CBL0137 leads to the reduction in the number of transformed foci in Ifnar-dependent manner. MEF of different background were transduced with lentiviral HrasV12 followed by treatment with 250 nM CBL0137 for 48 hr. Number of foci was detected on day 10 after transduction. Cultures from two embryos of each genotype were used. Mean of two replicates ± SD. * - p<0.05. B-C. Effect of CBL0137 on the growth of wild type (wt) (B) of Ifnar1-/- (C) transformed MEF in Ifnar1-/- mice. Group of five mice of each genotype were treated with vehicle of 70 mg/kg of CBL0137 once a week for 3 weeks. Mean tumor volume + SD. Asterisk – p<0.05 between CBL0137 and vehicle-treated groups.
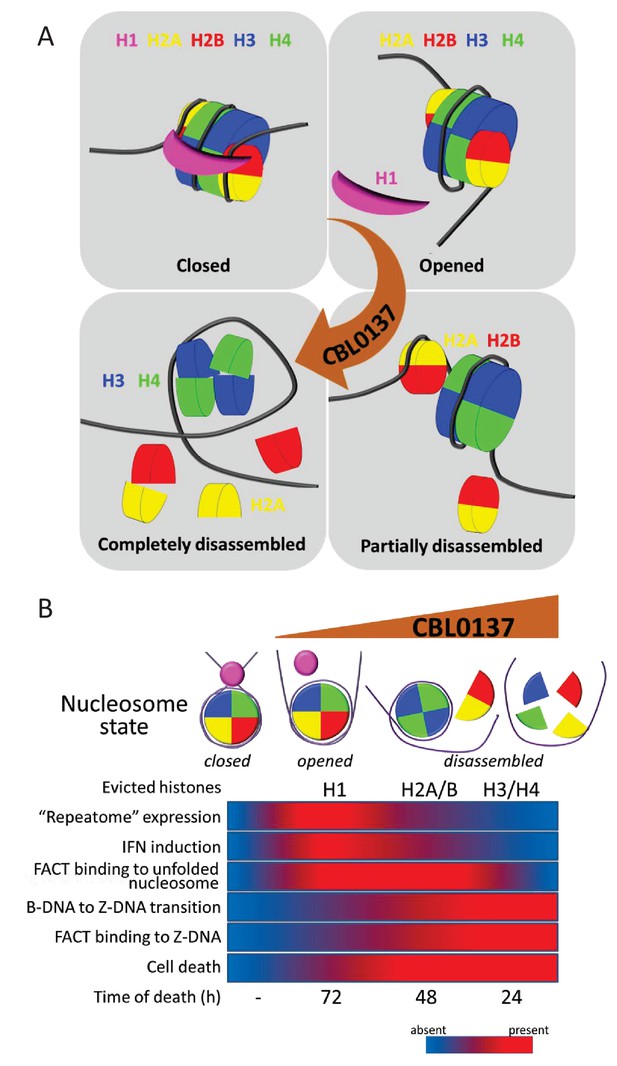
Proposed scheme of nucleosome states in the absence and presence of increasing concentrations of CBL0137 (A) and associated biological processes in cells (B).
https://doi.org/10.7554/eLife.30842.025Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30842.026





