Intrinsic disorder within AKAP79 fine-tunes anchored phosphatase activity toward substrates and drug sensitivity
Figures
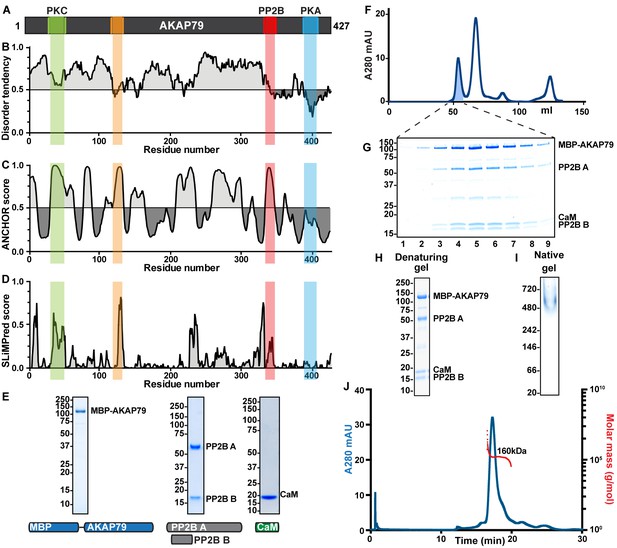
Characterizing disorder and short linear motifs in AKAP79 complexes.
(A) Primary topology of AKAP79, with new and known binding sites notated and shaded. (B) IUPred prediction of disordered regions of AKAP79. (C) PONDR prediction of disordered regions of AKAP79 (D) ANCHOR prediction of short linear motifs in AKAP79. (E) SLiMPred prediction of short linear motifs in AKAP79. (F) SDS-PAGE gels and constructs used for individual subunits of an MBP-AKAP79/PP2B/CaM complex. (G) Gel filtration of a fully assembled AKAP79/PP2B/CaM complex. (H). SDS-PAGE gel of fractions pooled for further analysis. (I) SEC-MALS of MBP-AKAP79/PP2B/CaM. A280 in blue, molecular weight measurement in red. Insets: denaturing and native gels of the protein complex.
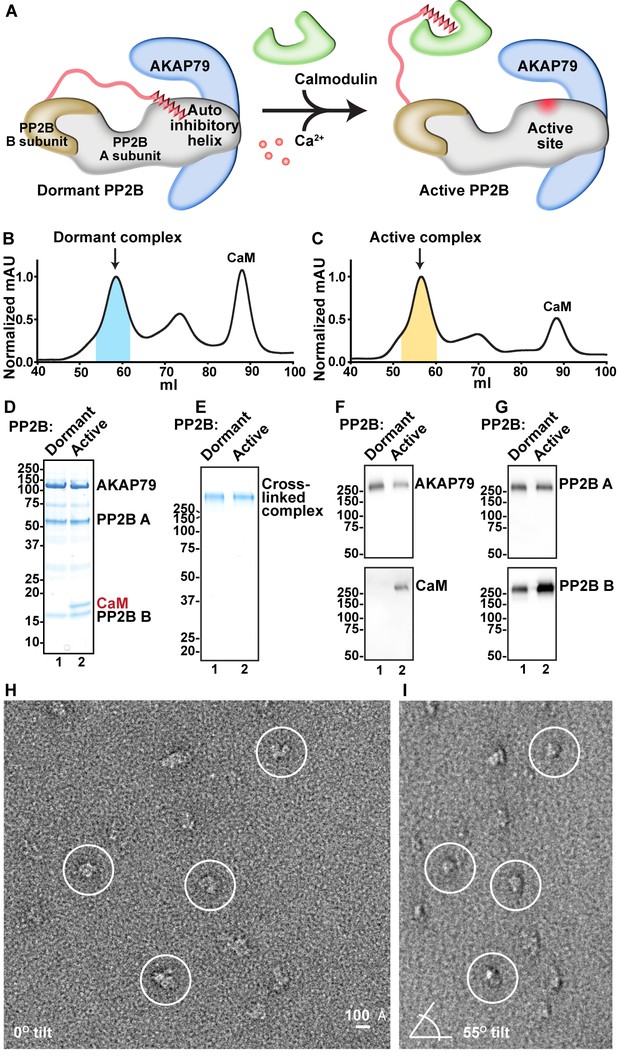
Analysis of compositional and conformational changes induced by Ca2+.
(A) Simplified schematic of general PP2B structural rearrangements upon addition of Ca2+/CaM. (B) SEC UV trace of AKAP79/PP2B/CaM complex obtained in the absence of Ca2+. Blue shading indicates fractions analyzed. (C) SEC UV trace of AKAP79/PP2B/CaM complex obtained in the presence of Ca2+. Yellow shading indicates fractions analyzed. (D) SDS-PAGE of pooled peak fractions from dormant and active complexes. (E) SDS-PAGE of GraFix purification and crosslinking of dormant and active complexes. (F) Western blots for the C-terminal region of AKAP79 and CaM. (G) Western blots for PP2B A and B subunits. (H) Example untilted micrograph of the dormant complex. (I) Example tilted micrograph of the dormant complex. Paired particles indicated by circles.
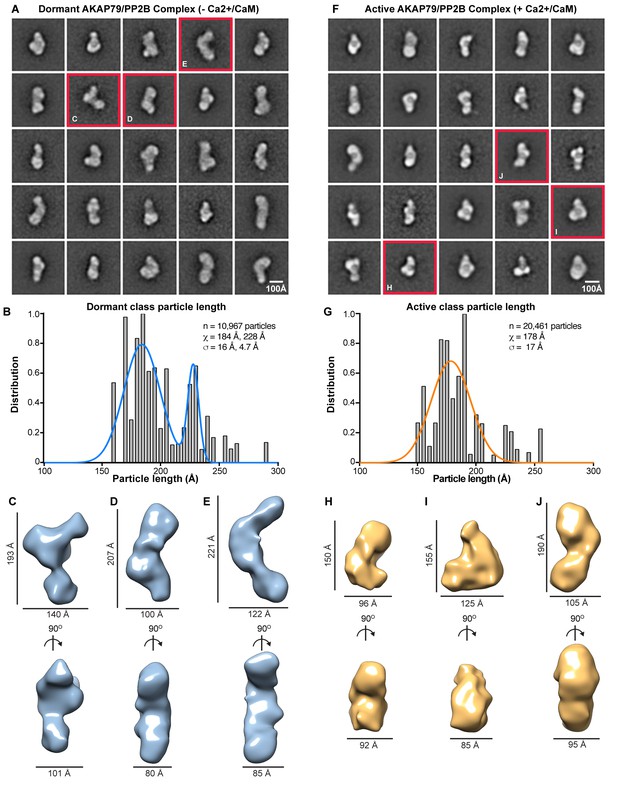
EM analysis of AKAP79/PP2B complexes.
(A) Reference-based class averages of the dormant complex. (B) Histogram analysis of dormant class dimensions. Bimodal Gaussian fit in blue. (C–E) Sample 3-D models of dormant AKAP79/PP2B illustrating representative states of the complex, notated with dimensions. (F) Reference-based class averages of active AKAP79/PP2B/CaM. (G) Histogram analysis of active class dimensions. Gaussian fit in orange. (H–J) Sample 3-D models of active AKAP79/PP2B/CaM, annotated with dimensions.
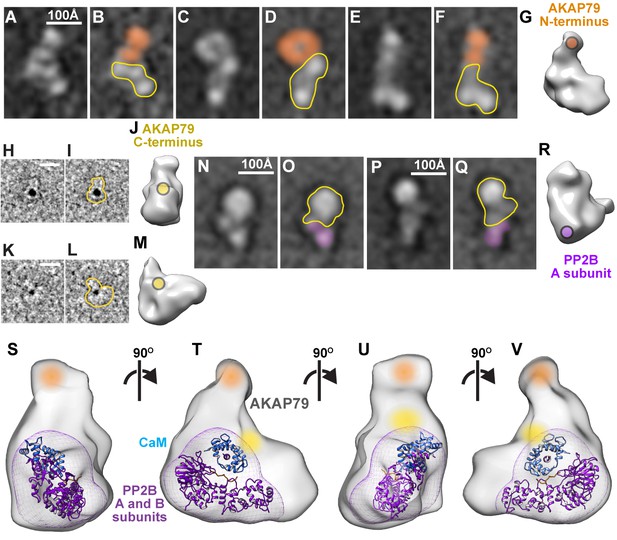
Subunit mapping of AKAP79/PP2B/CaM complexes.
(A–F) Reference-free class averages of AKAP79/PP2B/CaM assemblies complexed with anti-MBP-AKAP79 Fab fragments. Density corresponding to anti-MBP Fab fragment notated with orange. AKAP79/PP2B/CaM complex outlined in yellow (G) 3-dimensional model with anti-MBP Fab binding site indicated in orange. (H, K) Representative Ni-NTA gold-labeled single particle images of AKAP79/PP2B/CaM, 10 Å low-pass filtered for visualization. (I, L) Gold-labeled particle outlines indicated in yellow. (J, M) 3-dimensional models with gold-labeled 10x-His tag indicated in yellow. (N–Q) Reference-free class averages of AKAP79/PP2B/CaM assemblies complexed with anti-Flag-PP2B Fab fragments. Density corresponding to anti-MBP Fab fragment denoted with purple. AKAP79/PP2B/CaM complex outlined in yellow (R) 3-dimensional model with anti-Flag-PP2B Fab binding site indicated in purple. (S–V) 3-dimensional model of active AKAP79/PP2B/CaM complex with subunit labels notated in color and crystal structures of a PP2B (purple)/CaM (blue, PDB: 1YR5) complex docked within the density, views rotated successively around a vertical axis by 90˚. See also Figure 4—video 1.
3D RCT model with subunit crystal structures placed within EM model based on labeling experiments.
https://doi.org/10.7554/eLife.30872.007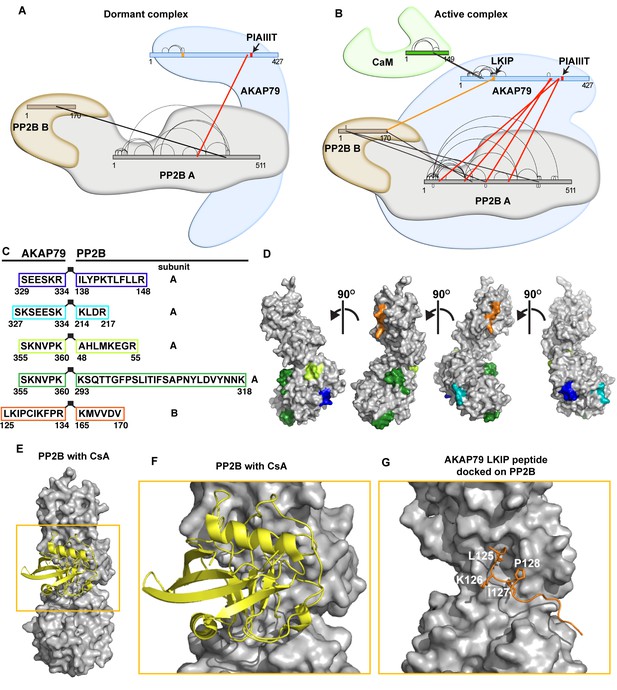
Crosslinking/mass-spectrometry of AKAP79/PP2B interactions.
(A) Crosslink map of residues observed in dormant complexes. Thick lines indicate intermolecular crosslinks, and red lines indicate crosslinks from PP2B to AKAP79. See also Supplementary file 1. (B) Crosslink map of residues that were observed in active complexes conditions. Thick lines indicate intermolecular crosslinks, and red lines indicate crosslinks from PP2B to AKAP79. Orange line indicates selected crosslink from AKAP79-K126 to PP2B-K165. (C) Table of AKAP79-PP2B crosslinked peptides observed in the active complex, color-coded. (D) Cognate binding surfaces for these peptides mapped onto the PP2B surface (gray), color-coded with the same scheme as the previous panel (PDB: 5SVE). (E) PP2B (gray) with cyclosporin/cyclophilin complex (yellow) (PDB: 1MF8). (F) Inset showing the immunophilin binding site. (G) Rosetta FlexPepDock model of AKAP79 crosslinked peptide (orange) complexed with PP2B (gray).
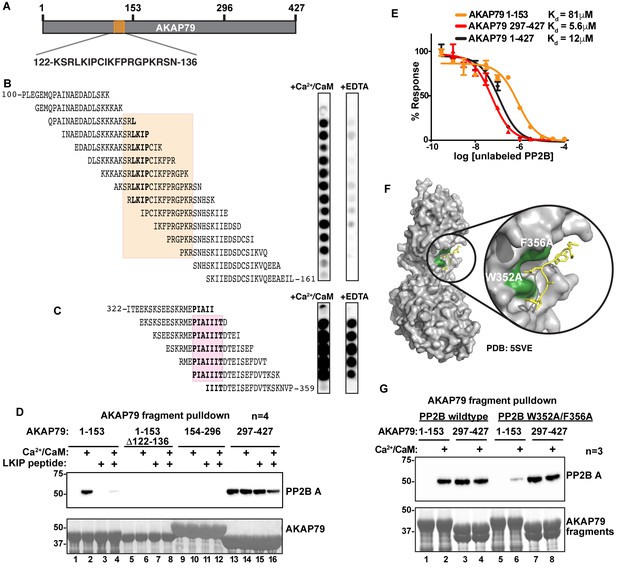
Mapping and characterization of the LKIP binding interfaces for AKAP79 and PP2B.
(A) Fragments of AKAP79 used in subsequent experiments. (B) PP2B overlay of synthesized peptides in the LKIP region ± Ca2+/CaM. Shading indicates sequences shared by Ca2+/CaM-dependent binding peptides. (C) PP2B overlay of synthesized peptides in the PIAIIIT region ± Ca2+/CaM. Shading indicates sequences shared PP2B-binding peptides. (D) GST-AKAP79 fragment pulldowns of Flag-PP2B in the presence of Ca2+/CaM, and competition with 200 μM NFAT peptide. Lanes 5–8 show that deletion of AKAP79 122–136 abolishes binding. See also Figure 6—figure supplement 1. (E) AlphaScreen competition assay to calculate Kd values for interactions between PP2B and full-length AKAP79 (green), or fragments of AKAP 79 (C – blue, N – orange). Data are represented as mean ±SEM. (F) Structural model showing PP2B (gray) with the mutated LxVP binding pocket residues (green) interacting with an LxVP motif (yellow) (PDB: 5SVE). (G) Mutations were made to PP2B that are predicted to abolish the LxVP binding pocket. GST-pulldown assays were used to test mutant PP2B binding to the N-terminal fragment of AKAP79.
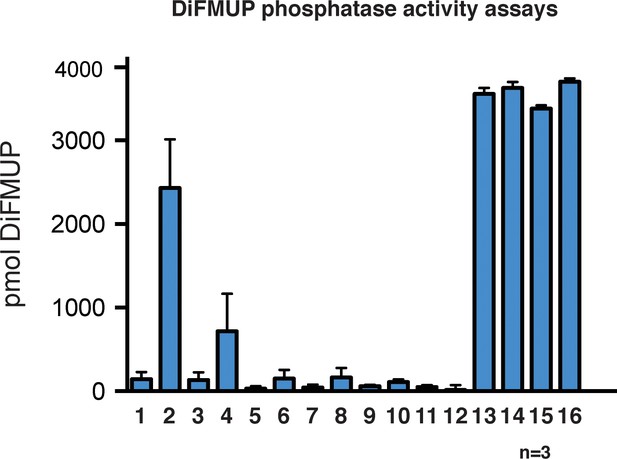
DiFMUP phosphatase activity assay.
Levels of dephosphorylated DiFMUP from corresponding samples in Figure 6D. Data are represented as mean ±SEM. N = 3.
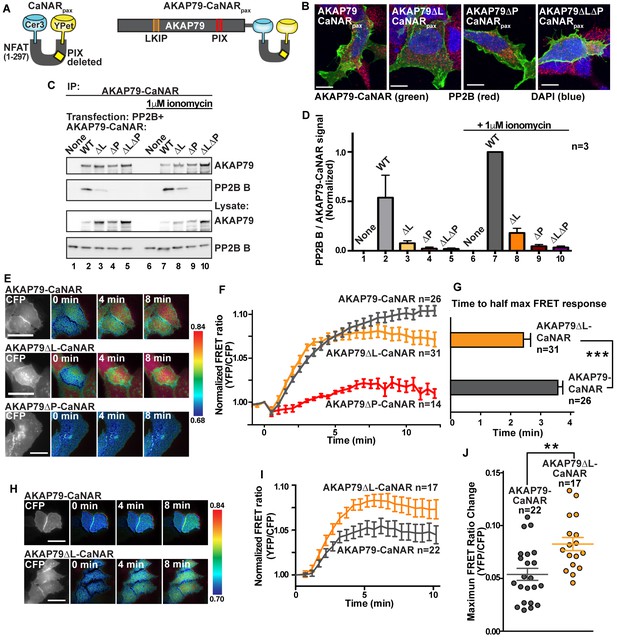
Fine tuning of PP2B sensitivity towards physiological ranges of calcium by AKAP79.
(A) Schematic of the CaNAR reporter with an ablated PxIxIT motif, fused in tandem with full-length AKAP79. (B) Confocal images of AKAP79-CaNAR variants (green), PP2B (red) and DAPI (blue) showing proper plasma membrane localization of various mutants. (C). Co-immunoprecipitation of PP2B with AKAP79-CaNAR variants under basal (lanes 1–5) and ionomycin treated (lanes 6–10) conditions. (D) Quantification of western blot signals of the ratio of co-immunoprecipitated PP2B B subunit to AKAP79-CaNAR variants, normalized to lane 7. Data are represented as mean ±SEM. (E) Representative frames of FRET movies comparing wild-type AKAP79-CaNAR with AKAP79∆L-CaNAR and AKAP79∆P-CaNAR signals. Colors are representative of the ratio value as described by the key to the right of each row. See also Figure 7—videos 1–3. (F) Time course of FRET ratio signal upon stimulation with 1 μM ionomycin at t = 0. (G) Time to half-max FRET signal after treatment with 1 μM ionomycin. Data are represented as mean ±SEM. (*** indicates p<0.001, unpaired t-test). (H) Representative frames of FRET movies comparing wild-type AKAP79-CaNAR with AKAP79∆L-CaNAR signals after treatment with lower ionomycin concentrations (100 nM). See also Figure 7—videos 4 and 5. (I) Time course of FRET ratio signal upon stimulation with 100 nM ionomycin at t = 0. (J) Treatment of cells with 100 nM ionomycin reveals differences in the max response of each variant. Data are represented as mean ±SEM. (** indicates p<0.01, unpaired t-test). See also Figure 7—figure supplement 1.
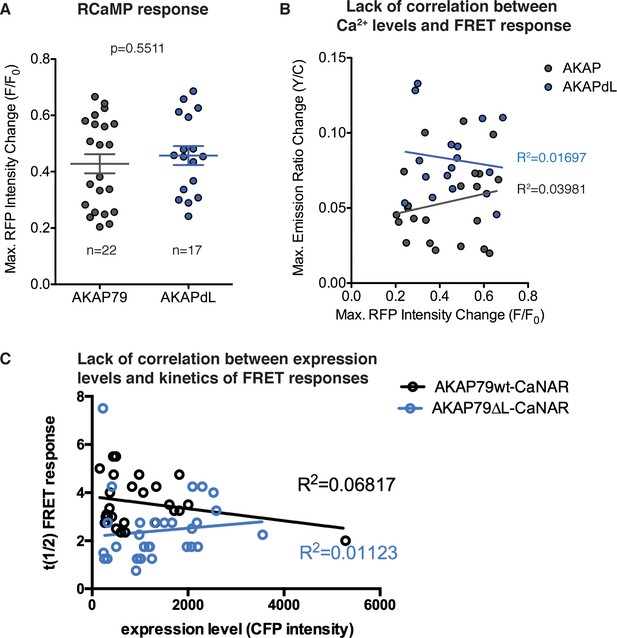
RCaMP measurements of calcium.
(A) Calcium levels were measured after ionomycin treatment (100 nM) to ensure that stimulation did not significantly vary between AKAP79-CaNAR (n = 22) and AKAP79∆L-CaNAR (n = 17). Data are represented as mean ±SEM. (B). Analysis of correlation shows that variation in calcium levels do not correlate with changes in CaNAR FRET responses. (C) The kinetics of FRET responses (t1/2) do not depend on expression levels of the CaNAR reporters, as measured by CFP intensity.
Representative response of wild-type AKAP79-CaNAR to 1 μM ionomycin.
https://doi.org/10.7554/eLife.30872.013Representative response of AKAP79∆L-CaNAR to 1 μM ionomycin.
https://doi.org/10.7554/eLife.30872.014Representative response of AKAP79∆P-CaNAR to 1 μM ionomycin.
https://doi.org/10.7554/eLife.30872.015Representative response of wild-type AKAP79-CaNAR to 100 nM ionomycin.
https://doi.org/10.7554/eLife.30872.016Representative response of wild-type AKAP79∆L-CaNAR to 100 nM ionomycin.
https://doi.org/10.7554/eLife.30872.017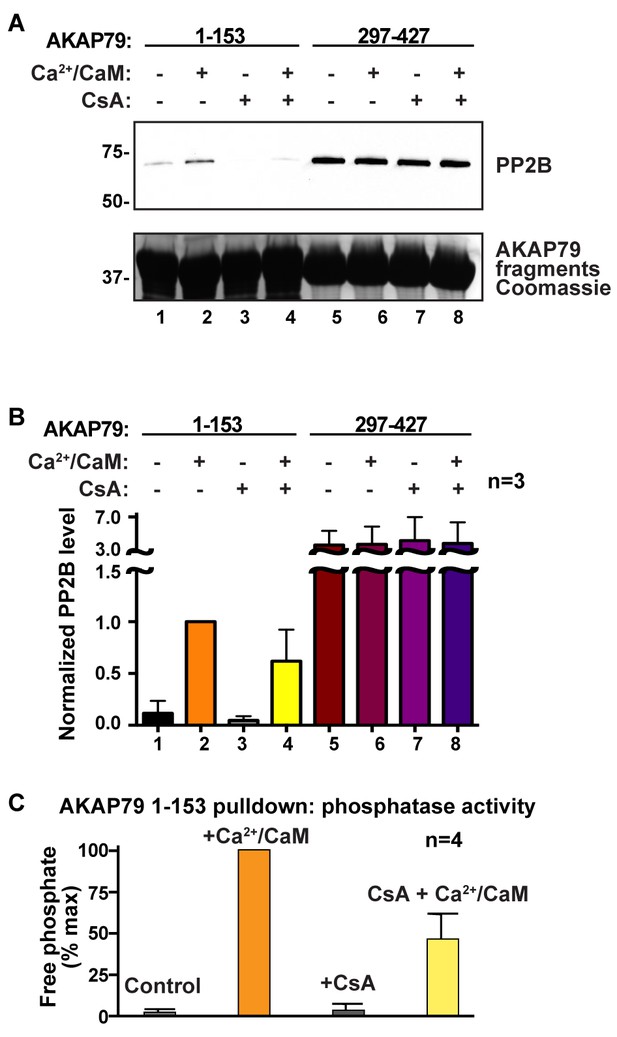
AKAP79 and cyclosporin bind to a common surface on PP2B.
(A) GST-pulldown experiments using N- and C- fragments of AKAP79 and testing for competition using cyclosporin/cyclophilin complexes, n = 3. (B) Quantification of PP2B signals in GST-pulldowns, normalized to lane 2, n = 3. Data are represented as mean ± SEM. (C) Phosphatase activity assay on samples from lanes 1–4 using a phosphopeptide substrate, n = 4. Data are represented as mean ± SEM.
Additional files
-
Supplementary file 1
(Related to Figure 5) Complete table of non-redundant observed crosslinked peptides in the presence of Ca2+ or EDTA.
- https://doi.org/10.7554/eLife.30872.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30872.020





