Ragulator and GATOR1 complexes promote fission yeast growth by attenuating TOR complex 1 through Rag GTPases
Figures
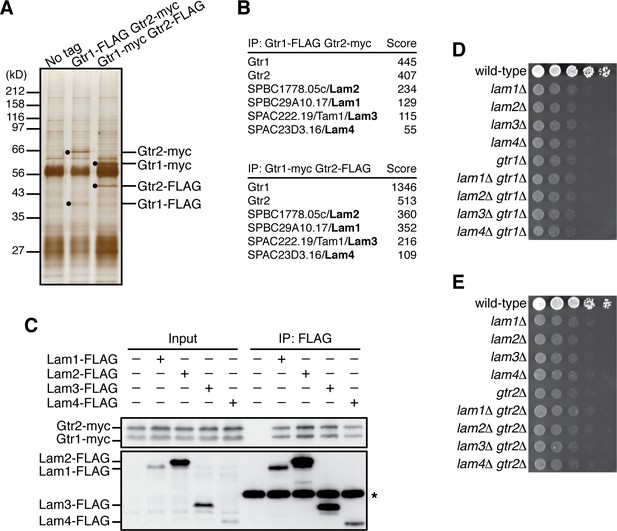
Identification of the Lam proteins that interact with the Gtr1-Gtr2 Rag GTPase heterodimer.
(A) Affinity-purification of the Gtr1-Gtr2 heterodimer from S. pombe. The heterodimer was purified from the cell lysate of gtr1:FLAG gtr2:myc and gtr1:myc gtr2:FLAG strains by two successive immunoprecipitation procedures using anti-FLAG and anti-myc antibodies, and resolved on SDS-PAGE followed by silver staining. A wild-type strain that expresses the untagged Gtr proteins (‘No tag’) was used as a negative control. The protein bands corresponding to the tagged Gtr1 and Gtr2 are indicated by black dots. (B) Mass spectrometric analyses of proteins co-purified with the Gtr1-Gtr2 heterodimer in the experiments shown in (A). Two or more peptides were identified for each protein listed in the tables. The sum of peptide scores that exceed the 95% confidence level (p<0.05) is also shown for each of the identified proteins. (C) Confirmed physical interactions between the Lam proteins and Gtr1-Gtr2. Crude lysate (‘Input’) was prepared from gtr1:myc gtr2:myc strains expressing one of the Lam proteins with the FLAG tag, and anti-FLAG immunoprecipitates (‘IP:FLAG’) were analyzed by immunoblotting. Immunoglobulin in the immunoprecipitates is asterisked. (D–E) The lam and gtr genes function in the same pathway. The indicated single and double mutants as well as a wild-type strain were grown in EMM liquid medium and their serial dilutions were spotted onto YES agar medium for a growth assay at 30˚C.
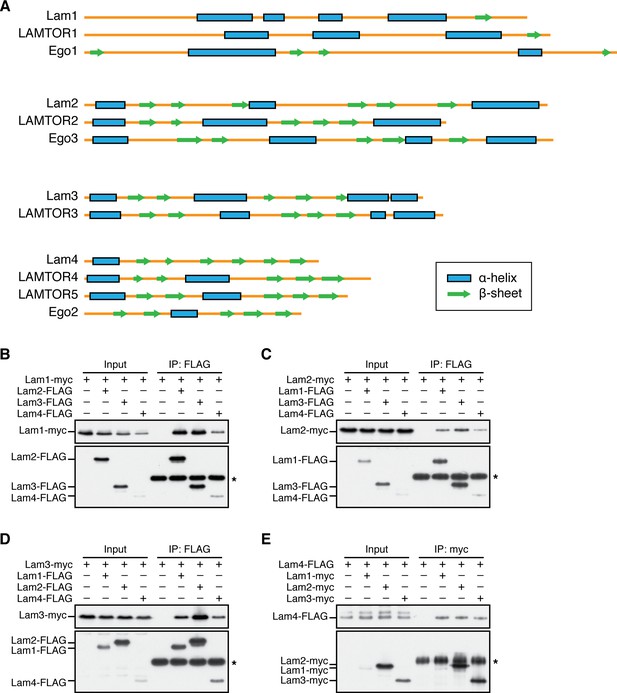
The S. pombe Lam proteins form a complex equivalent of mammalian Ragulator and the Ego ternary complex in budding yeast.
(A) The secondary structures of the Lam proteins in S. pombe, human Ragulator subunits LAMTOR1 ~5 and the budding yeast Ego complex components are predicted using the Jpred server (http://www.compbio.dundee.ac.uk/jpred/). Boxes and arrows indicate α-helix and β-sheet structures, respectively. (B–E) Confirmed pairwise interactions among the Lam proteins. lam1:myc (B), lam2:myc (C), lam3:myc (D) and lam4:FLAG (E) strains expressing one of the other Lam proteins with a different epitope tag were analyzed by anti-FLAG or anti-myc immunoprecipitation followed by immunoblotting. Immunoglobulin in the immunoprecipitates is asterisked.
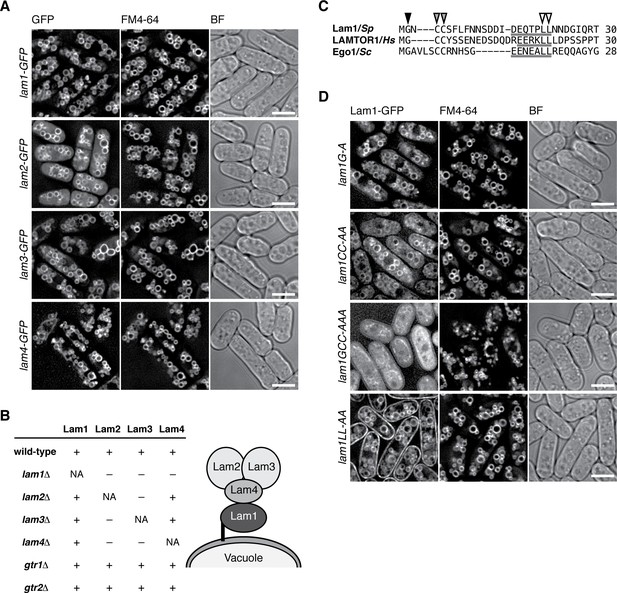
The Lam protein complex is localized to vacuolar membranes.
(A) The chromosomal lam genes were tagged with the GFP sequence and the strains were grown in EMM at 30˚C for microscopy, with vacuolar membranes visualized by the fluorescent dye FM4-64. Z-axial images were collected and mid-section images after deconvolution are shown. BF, bright-field image. Bars, 5 µm. (B) Interdependent vacuolar localization of the Lam proteins. See Figures 2A and Figure 2—figure supplements 1 and 2 for representative microscopy images. ‘+’, localization to vacuolar membranes; ‘–’, defused in the cytosol. Predicted architecture of the Lam complex on the vacuolar surface is also shown as a schematic diagram. (C) Sequence alignment of the N-terminal regions of S. pombe Lam1 (UniProtKB ID: C6Y4C6), human LAMTOR1 (Q6IAA8) and Ego1 in Saccharomyces cerevisiae (Q02205). The conserved Gly (black arrowhead) and Cys (gray arrowheads) residues are potential sites for myristoylation and palmitoylation, respectively. Putative vacuole/lysosome localization signal sequences composed of acidic residues followed by a di-leucine motif (open arrowheads) are underlined. (D) The sequence motifs shown in (C) are important for the vacuolar localization of Lam1. The lam1:GFP strains with alanine-substitutions at the potential myristoylation site (G–A), palmitoylation sites (CC-AA), both of them (GCC-AAA) or the di-leucine (LL-AA) motif were analyzed as in (A). Bars, 5 µm.
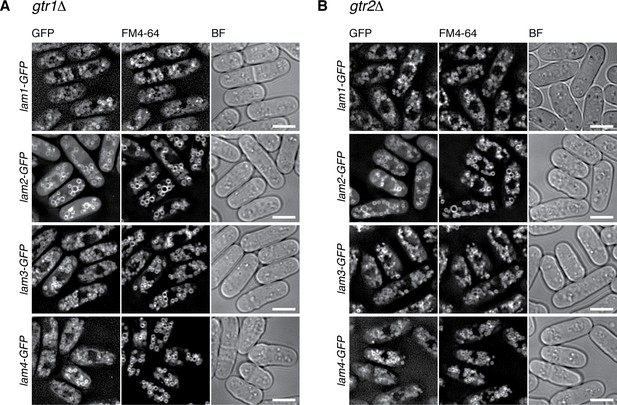
Vacuolar localization of the Lam proteins does not require Gtr1-Gtr2.
The chromosomal lam genes were tagged with the GFP sequence in the gtr1∆ (A) and gtr2∆ (B) strains, which were grown in EMM at 30 ˚C for microscopy, with vacuolar membranes visualized by the fluorescent dye FM4-64. z-axial images were collected and mid-section images after deconvolution are shown. BF, bright-field images. Bars, 5 µm.
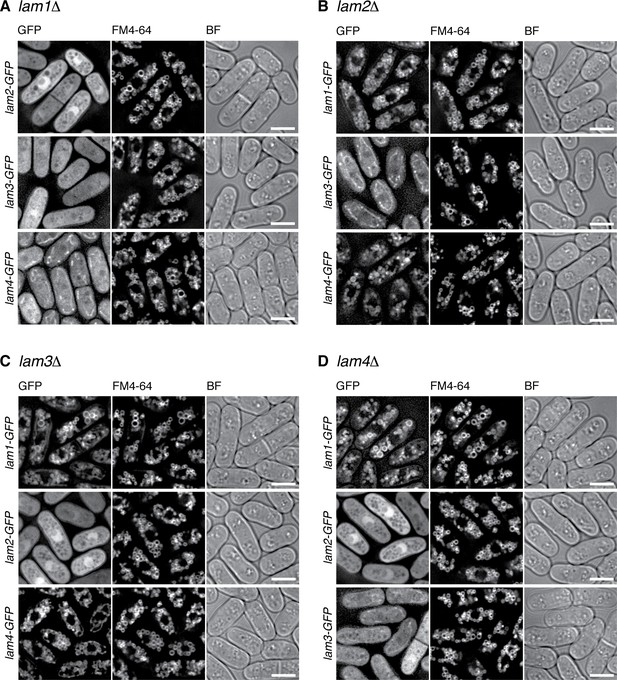
Interdependent vacuolar localization of the Lam proteins.
(A) Lam1 tethers the other Lam proteins to the vacuole. The lam1∆ mutant strains expressing GFP-tagged Lam2, Lam3 or Lam4 from their chromosomal loci were grown in EMM at 30 ˚C for microscopy, with vacuolar membranes visualized by the fluorescent dye FM4-64. z-axial images were collected and mid-section images after deconvolution are shown. BF, bright-field images. Bars, 5 µm. (B) The lam2∆ mutant strains expressing GFP-tagged Lam1, Lam3 or Lam4 were analyzed as in (A). (C) The lam3∆ mutant strains expressing GFP-tagged Lam1, Lam2 or Lam4 were analyzed as in (A). (D) The lam4∆ mutant strains expressing GFP-tagged Lam1, Lam2 or Lam3 were analyzed as in (A).
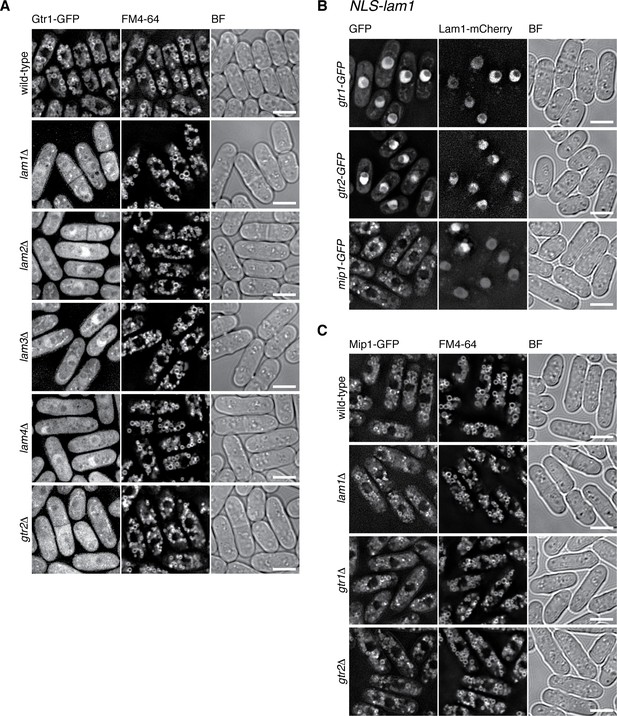
The Lam complex is the Ragulator equivalent that tethers Gtr1-Gtr2 to vacuolar membranes.
(A) The Lam proteins and Gtr2 are essential for the vacuolar localization of Gtr1. Wild-type and the indicated mutant strains expressing GFP-tagged Gtr1 from its chromosomal locus were analyzed by fluorescence microscopy as in Figure 2A. Bars, 5 µm. (B) Artificial nuclear targeting of Lam1 translocates Gtr1-Gtr2, but not TORC1, to the nucleus. Lam1GCC-AAA that cannot localize to vacuoles (Figure 2D) was fused to the SV40 nuclear localization signal (NLS) and mCherry sequences, and expressed in the gtr1:GFP, gtr2:GFP and mip1:GFP strains for fluorescence microscopy. Bars, 5 µm. (C) The Ragulator-Rag complex is not required for vacuolar localization of TORC1. Wild-type and the indicated mutant strains that express the Mip1 subunit of TORC1 with the GFP tag were analyzed by fluorescence microscopy. Bars, 5 µm.
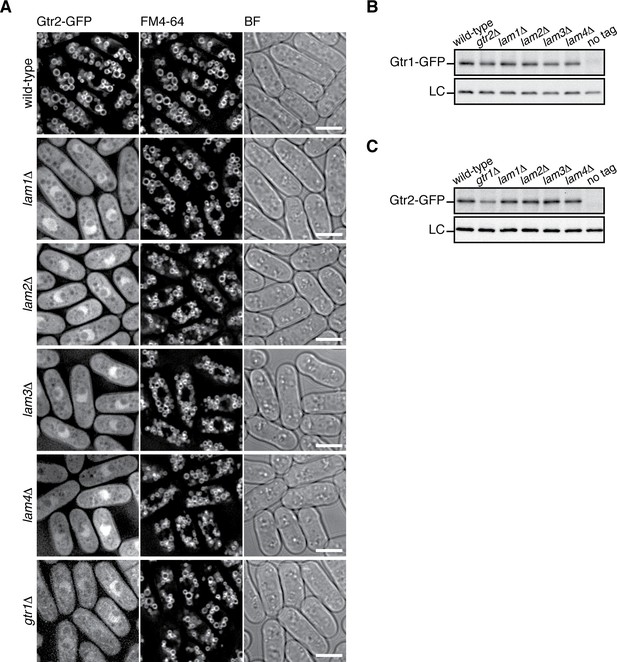
The Lam complex is important for the vacuolar localization, but not the stability, of the Gtr1-Gtr2 proteins.
(A) The Lam proteins and Gtr1 are essential for the vacuolar localization of Gtr2. Wild-type and the indicated mutant strains expressing GFP-tagged Gtr2 from its chromosomal locus were analyzed by fluorescence microscopy as in Figure 2A. BF, bright-field images. Bars, 5 µm. (B–C) Loss of Ragulator does not significantly affect the stability of Gtr1 and Gtr2. Wild-type and the indicated mutant strains carrying the gtr1:GFP (B) or gtr2:GFP (C) loci were grown in EMM at 30 ˚C and their lysate was analyzed by anti-GFP immunoblotting. Anti-Spc1 MAPK immunoblotting served as a loading control (LC).
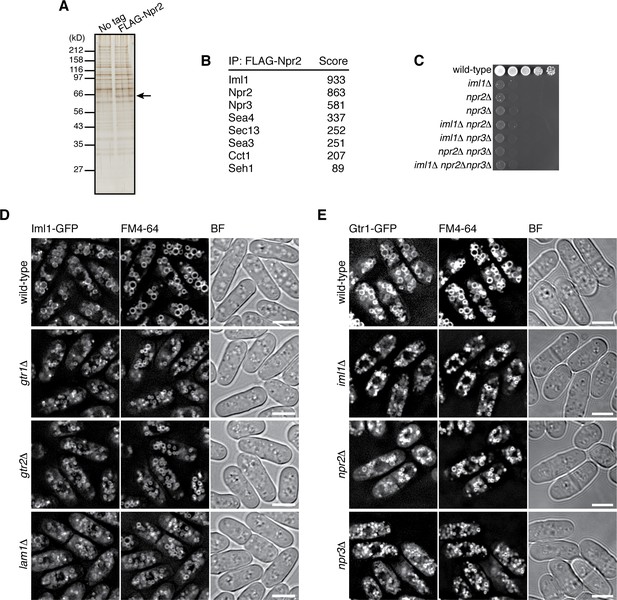
Identification of the fission yeast GATOR1 complex.
(A) Affinity purification of FLAG-tagged Npr2 and its associating proteins. Cells expressing FLAG-Npr2 were grown in YES medium and their lysate was subjected to anti-FLAG immunoprecipitation. The eluates were resolved on SDS-PAGE followed by silver staining. A wild-type strain that expresses untagged Npr2 (‘No tag’) was used as a negative control. The protein band corresponding to FLAG-Npr2 is indicated by an arrow. (B) Mass spectrometric analyses of proteins co-purified with FLAG-Npr2 in the experiment shown in (A) identified S. pombe orthologs of the GATOR1 (Iml1 and Npr3) and GATOR2 (Seh1, Sea3, Sea4 and Sec13) subunits. Two or more peptides were identified for each protein listed in the table. The sum of peptide scores that exceed the 95% confidence level (p<0.05) is also shown for each of the identified proteins. (C) Growth defects of the mutants lacking the GATOR1 subunits. The indicated single, double and triple mutants as well as a wild-type strain were grown in EMM liquid medium and their serial dilutions were spotted onto YES agar medium at 30˚C. (D) Vacuolar localization of GATOR1 is independent of the Ragulator-Rag complex. The iml1:GFP locus was introduced to the wild type and the mutants lacking Gtr1-Gtr2 or Lam1 and the localization of Iml1-GFP was analyzed by fluorescence microscopy. Bars, 5 µm. (E) Vacuolar localization of the Gtr1 GTPase is independent of GATOR1. Wild-type and GATOR1 mutant strains expressing GFP-tagged Gtr1 from its chromosomal locus were analyzed by fluorescence microscopy. Bars, 5 µm.

Fission yeast Iml1 and Npr3 are orthologous to the mammalian GATOR1 components, DEPDC5 and Nprl3, respectively.
The amino acid sequences of (A) human DEPDC5 (UniProtKB ID: O75140-10) and S. pombe Iml1 (O74788), and (B) those of human Nprl3 (Q12980) and S. pombe Npr3 (Q9HGM7) were aligned using the CLUSTALW program (http://www.genome.jp/tools/clustalw/). Asterisks, identical amino acids; single and double dots, weakly and strongly similar amino acids, respectively. The ‘arginine finger’ conserved within the GAP domain of Iml1 orthologs (R854 in S. pombe Iml1) is highlighted in (A).
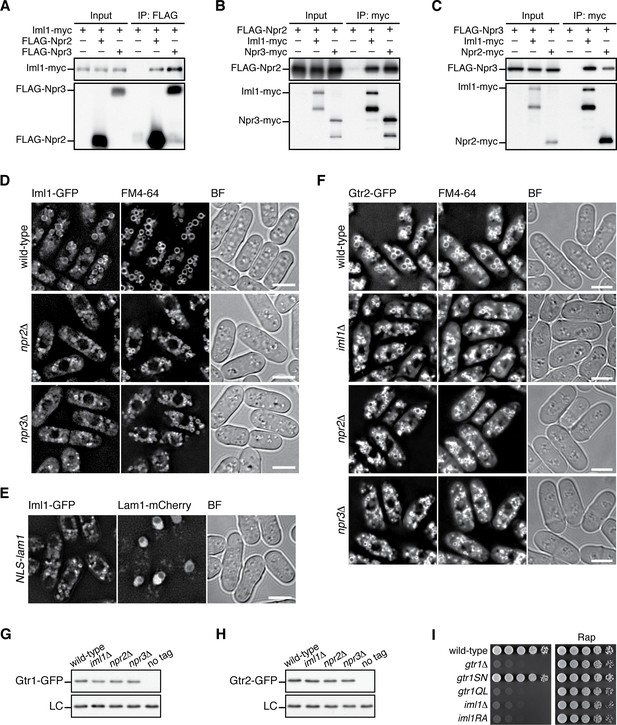
Expression and vacuolar localization of Gtr1-Gtr2 are not regulated by GATOR1.
(A-C) Confirmed pairwise interactions among the GATOR1 subunits. Crude lysate (“input”) was prepared from iml1:myc (A), FLAG:npr2 (B) and FLAG:npr3 (C) strains expressing one of the other GATOR1 subunits with a different epitope tag and analyzed by immunoprecipitation (“IP”) followed by immunoblotting.(D) Vacuolar localization of Iml1 does not require the other GATOR1 components, Npr2 and Npr3. The iml1:GFP locus was introduced to wild-type, npr2∆ and npr3∆ strains, and the cells grown in EMM at 30˚C were observed after vacuolar staining with the FM4-64 dye. Mid-section images are presented after deconvolution. BF, bright-field images. Bars, 5 µm.(E) Vacuolar localization of GATOR1 is independent of the Ragulator-Rag complex. The iml1:GFP locus was introduced to the strain with nuclear-targeted Lam1 (Fig. 3B), and the localization of Iml1-GFP was analyzed by fluorescence microscopy. (F) Vacuolar localization of Gtr2 is independent of GATOR1. Wild-type and GATOR1 mutant strains expressing GFP-tagged Gtr2 from its chromosomal locus were analyzed as in (D). Bars, 5 µm.(G-H) Loss of GATOR1 does not significantly affect the stability of the Gtr1 and Gtr2 proteins. Wild-type and the indicated mutants strains carrying the gtr1:GFP (G) or gtr2:GFP (H) loci were grown in EMM at 30˚C and their lysate was analyzed by anti-GFP immunoblotting. Anti-Spc1 MAPK immunoblotting served as a loading control (LC).(I) The conserved “arginine finger” within the GAP domain of Iml1 is essential for the GATOR1 function. The iml1R854A mutant (“iml1RA”), in which Iml1 Arg854 is substituted with Ala, and the other indicated strains were grown in liquid EMM, and their growth was tested at 30˚C on YES agar with and without 100 ng/ml rapamycin (Rap). iml1R854A cells exhibited phenotypes indistinguishable from those of the gtr1∆, gtr1QL and iml1∆.
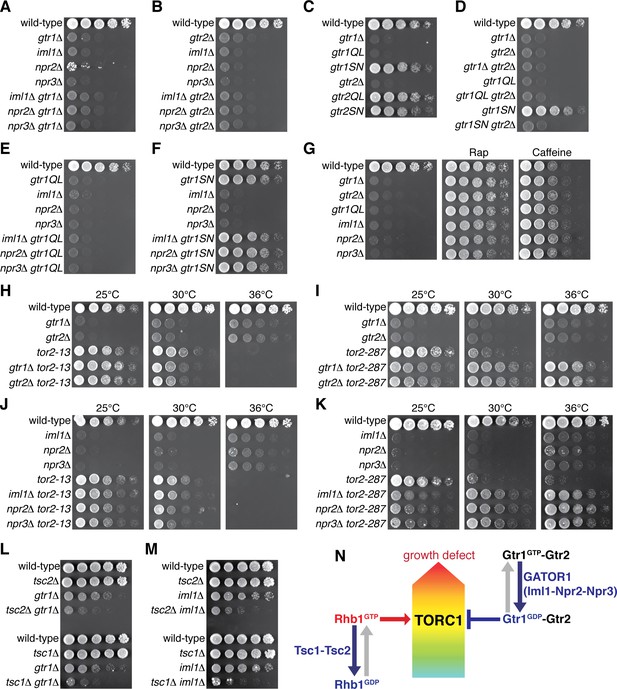
Gtr1GDP-Gtr2 attenuates TORC1 signaling for normal cell growth.
Wild-type and the indicated mutant strains were grown in EMM liquid medium and their serial dilutions were spotted onto YES agar media at 30˚C unless indicated otherwise. (A, B) GATOR1 and Gtr1-Gtr2 function in the same pathway. Growth of the indicated single and double mutants were compared. (C, D) Growth phenotypes of the strains expressing the GTP-locked (‘gtr1QL’ and ‘gtr2QL’) and GDP-locked (‘gtr1SN’ and ‘gtr2SN’) mutants of the Gtr GTPases. Wild-type and GDP-locked Gtr1 allow normal cell growth only in the presence of Gtr2. (E, F) Genetic interactions between GATOR1 and Gtr1-Gtr2. The growth defects caused by the loss of GATOR1 are complemented by expressing the GDP-bound Gtr1S16N. (G) TORC1 inhibitors suppress the growth defects of the mutants lacking Gtr1GDP-Gtr2. Growth of the indicated strains was tested on YES agar with 100 ng/ml rapamycin (Rap) or 10 mM caffeine. (H–K) Temperature-sensitive, hypomorphic mutations of the TOR kinase, tor2-13 and tor2-287, suppress the growth defects of the cells lacking Gtr1-Gtr2 or GATOR1. Growth of the indicated single and double mutants was tested at the permissive (25˚C), semi-permissive (30˚C) and restrictive (36˚C) temperatures of the tor2 mutants. (L–M) The GATOR1-Gtr pathway negatively regulates TORC1 independently of the Tsc complex. The growth defects of the gtr1∆ and iml1∆ mutants were accentuated in the absence of Tsc1 or Tsc2. (N) A working model for the TORC1 regulation by GATOR1 and Gtr1-Gtr2 in fission yeast. GATOR1 composed of Iml1, Npr2 and Npr3 functions as GAP for Gtr1, which forms heterodimer with Gtr2 to moderate TORC1 activation induced by the GTP-bound Rhb1 GTPase. Hyperactivation of TORC1 in the absence of Gtr1GDP-Gtr2 results in a growth defect as found in (A–K).
-
Figure 5—source data 1
Mutation sites used to generate the GDP-locked mutant forms of RagA/B GTPases.
Sequence alignment of the N-terminal regions of the RagA/B orthologs (UniProtKB IDs: Hs RagA, Q7L523; Hs RagB, Q5VZM2-1; Dm RagA, Q9VHJ4; Sc Gtr1, Q00582; Sp Gtr1, O74824) and that of K-Ras (P01116-1) by the CLUSTALW program (http://www.genome.jp/tools/clustalw/) is shown. Among the Ras-superfamily GTPases, the boxed Ser/Thr residues are conserved and their substitutions have been used to generate RagA/B and K-Ras mutants predicted to be restricted to the GDP-bound conformations in a number of studies including those listed in the table below. Gtr1 Ser20 mutated in the study by Ma et al. (2016) is marked by a red square.
- https://doi.org/10.7554/eLife.30880.014
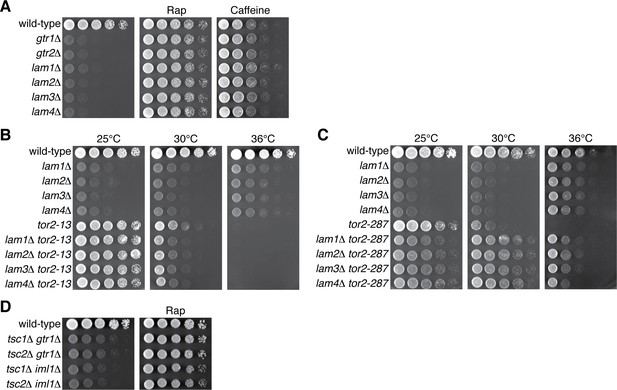
Ragulator is important for attenuation of TORC1 signaling in S. pombe.
Wild-type and the indicated mutant strains were grown in liquid EMM and their serial dilutions were spotted onto YES agar for a growth assay. (A) TORC1 inhibitors suppress the growth defects of the mutants lacking the Ragulator components Lam1 ~4. Growth of the indicated strains was tested at 30 ˚C on YES agar with 100 ng/ml rapamycin (Rap) or 10 mM caffeine. (B–C) Temperature-sensitive, hypomorphic mutations of the TOR kinase, tor2-13 and tor2-287, suppress the growth defects of the cells lacking the Ragulator components. Growth of the indicated single and double mutants was tested at the permissive (25 ˚C), semi-permissive (30 ˚C) and restrictive (36 ˚C) temperatures of the tor2 mutants. (D) Rapamycin suppresses the growth defects of the tsc∆ gtr1∆ and tsc∆ iml1∆ double mutants. Growth of the indicated strains was tested at 30 ˚C with and without 100 ng/ml rapamycin (Rap).
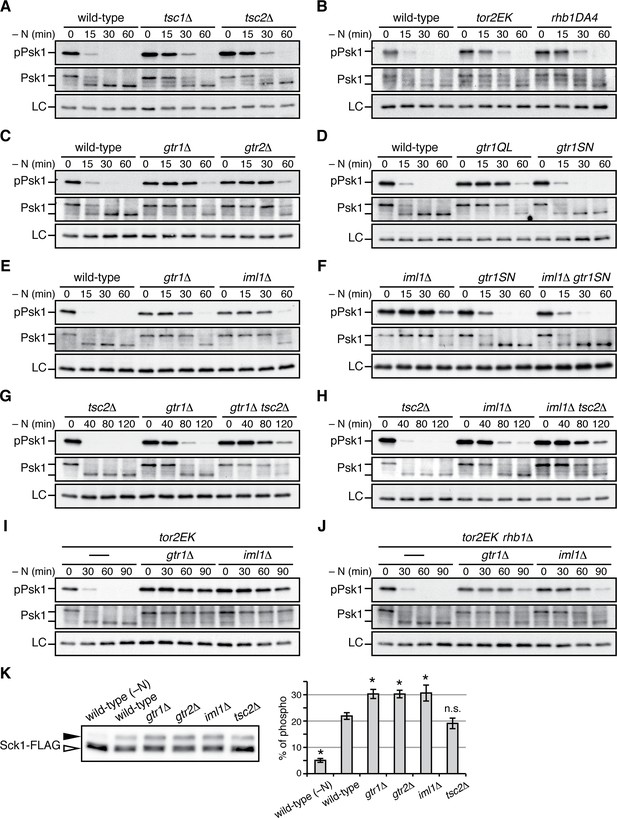
Gtr1GDP-Gtr2 negatively regulates TORC1 signaling.
(A–J) TORC1 activity was monitored by immunoblotting to detect the TORC1-dependent phosphorylation of Psk1 (‘pPsk1’), along the time course after the indicated strains exponentially growing in EMM at 30˚C were shifted to the same medium without nitrogen source. The samples were also probed with anti-Psk1 antibodies (‘Psk1’) as well as anti-Spc1 MAPK antibodies to control loading (‘LC’). Inactivation of TORC1 after nitrogen starvation was delayed in the strains lacking the Tsc complex (A) and those carrying the activating mutations tor2EK or rhb1DA4 (B). Similarly, loss of Gtr1-Gtr2 or GATOR1 (‘iml1∆') resulted in delayed starvation response (C, E), while the iml1∆ defect was complemented by expressing the GDP-locked mutant (‘gtr1SN’) form of Gtr1 (D, F). The gtr1∆ and iml1∆ mutations delayed the starvation response even in the tsc2∆ (G, H) and rhb1∆ (J) backgrounds, suggesting that Gtr1-Gtr2 and GATOR1 negatively regulate TORC1 signaling independently of the TSC-Rhb1 pathway. (K) TORC1-dependent phosphorylation of Sck1 is augmented in the absence of Gtr1GDP-Gtr2. The sck1:FLAG strains carrying the indicated mutations are grown in EMM at 30˚C and their lysate was analyzed by immunoblotting. As a negative control with inactive TORC1, a wild-type strain expressing Sck1-FLAG was starved for nitrogen for 1 hr (‘-N’). The band intensity of the phosphorylated (filled arrowhead) and unphosphorylated (open arrowhead) forms of Sck1-FLAG was quantified, and the percentages of the phosphorylated form to the total Sck1-FLAG level in each sample are presented in the bar graph as means ± SD (n = 4 independent experiments). *p<0.01; n.s., not significant, compared to the wild-type control using Student’s t-test.
-
Figure 6—source data 1
Source data for Figure 6K.
- https://doi.org/10.7554/eLife.30880.016
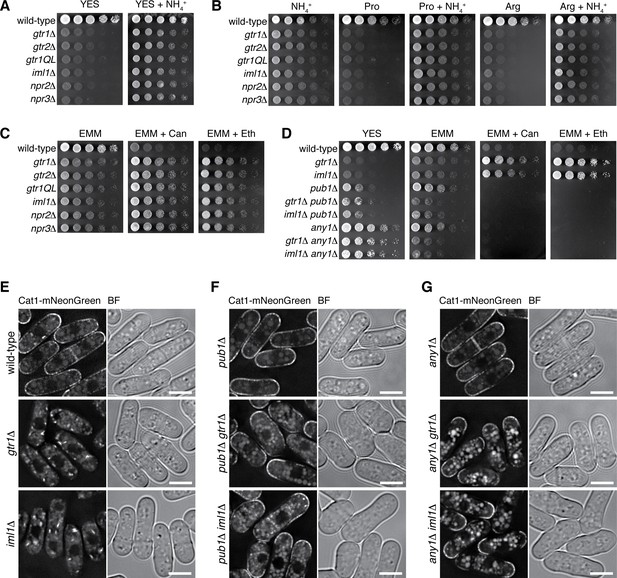
Loss of functional Gtr1GDP-Gtr2 results in a defect in amino acid uptake.
(A–D) The indicated strains were grown in EMM and their serial dilutions were spotted onto different agar media at 30˚C. The growth defects of the strains lacking Gtr1-Gtr2 or GATOR1 on yeast extract medium (YES) were partially complemented by supplementing the medium with 5 mg/ml NH4Cl (A). The mutants also showed severe growth defects on minimal media containing amino acid (20 mM Pro or Arg) as sole nitrogen source, while the phenotype was ameliorated by adding 5 mg/ml NH4Cl (B). In comparison to wild-type cells, those mutants were more resistant to canavanine (‘Can’, 60 µg/ml) and ethionine (‘Eth’, 30 µg/ml), toxic analogs of arginine and methionine, respectively (C), but the phenotype was reversed by inactivating the Pub1-Any1 ubiquitin ligase complex, which promotes internalization of amino acid transporters (D). (E–G) Translocation of the amino acid transporter Cat1 from the plasma membrane to the cytoplasm in the absence of Gtr1-Gtr2 or GATOR1. The chromosomal cat1+ gene was tagged with the mNeonGreen sequence in the indicated strains for microscopic analysis. The pub1∆ and any1∆ mutations restored the plasma membrane localization of Cat1 in the gtr1∆ and iml1∆ strains. Bars, 5 µm.
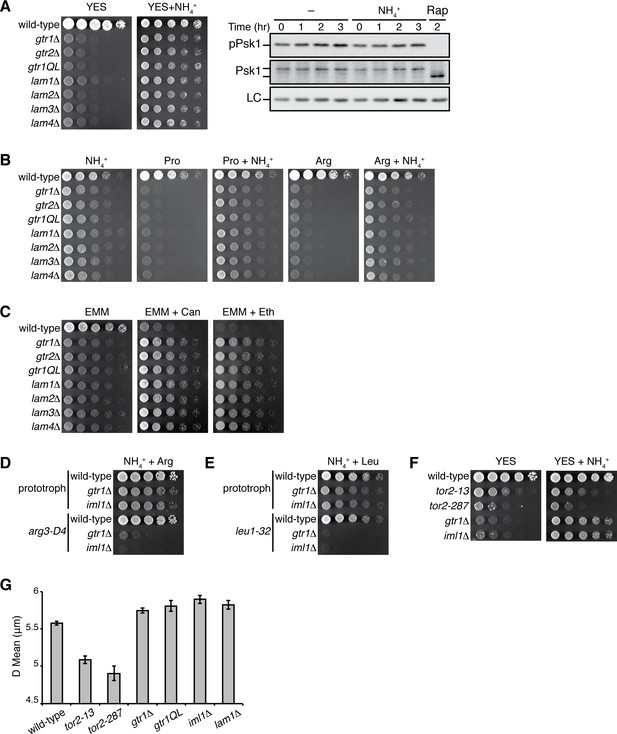
Loss of Gtr1GDP-Gtr2 or Ragulator results in a defect in amino-acid uptake.
(A) The growth defects of the strains lacking Gtr1GDP-Gtr2 or the Ragulator components Lam1 ~4 were partially complemented by supplementing the YES medium with 5 mg/ml NH4Cl. The indicated strains were grown in liquid EMM and their serial dilutions were spotted onto the indicated agar media at 30 ˚C. TORC1 activity was monitored by immunoblotting as in Figure 6 after wild-type cells growing in YES at 30°C were shifted to the same medium supplemented with NH4Cl or 200 ng/ml rapamycin (Rap) in order to confirm that NH4Cl does not significantly affect TORC1 activity in YES. (B–C) Those mutants also showed severe growth defects on minimal media containing amino acid (20 mM Pro or Arg) as sole nitrogen source, while the phenotype was ameliorated by adding 5 mg/ml NH4Cl (B). In comparison to wild-type cells, the mutants were more resistant to canavanine (‘Can’, 60 µg/ml) and ethionine (‘Eth’, 30 µg/ml), toxic analogs of arginine and methionine, respectively (C). (D–E) gtr1∆ and iml1∆ mutant strains with amino-acid auxotrophy show severe growth defects even in the presence of amino acids supplemented to the growth medium. The indicated prototrophic, arginine-auxotrophic (arg3-D4) and leucine-auxotrophic (leu1-32) strains were grown in liquid YES supplemented with 200 ng/ml rapamycin, washed with EMM liquid, and their serial dilutions were spotted onto EMM agar media with 225 mg/L arginine (D) or 250 mg/L leucine (E) at 30 ˚C. (F) Unlike the gtr1∆ and iml1∆ mutants, the growth defects of the tor2-13 and tor2-287 are not suppressed by NH4Cl in the growth medium. Growth of the indicated strains was tested as in (A). (G) In contrast to the tor2 hypomorphic mutants, cells lacking Gtr1GDP-Gtr2 or Ragulator are not smaller than wild-type cells. The indicated strains were grown in EMM at 30 ˚C, and a particle analyzer was used to measure their cell volume, which is presented as equivalent spherical diameter in the bar graph (means ±SD, n = 3).
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1G.
- https://doi.org/10.7554/eLife.30880.019
Additional files
-
Supplementary File 1
List of key resources used in this study.
- https://doi.org/10.7554/eLife.30880.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30880.021





