Crucial role for T cell-intrinsic IL-18R-MyD88 signaling in cognate immune response to intracellular parasite infection
Figures
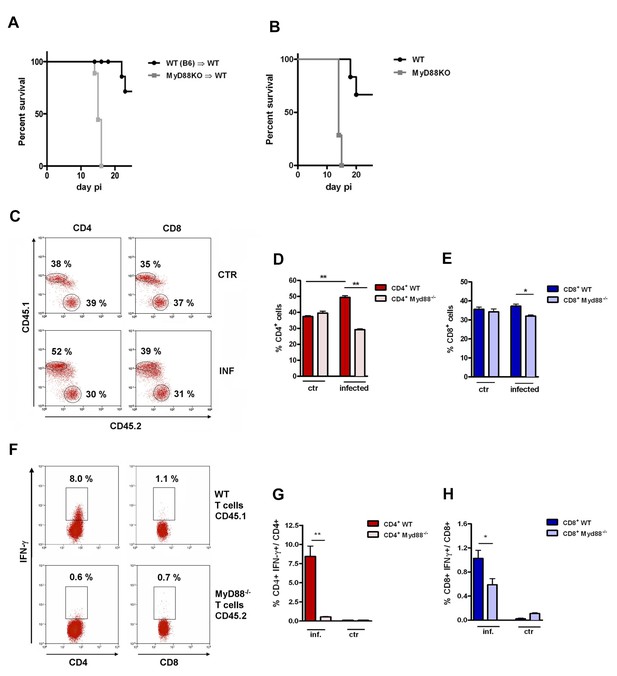
Lower expansion of IFN-γ+CD4+ Myd88-/- cells in infected mixed BM chimeras.
(A, B) Survival curves of mice infected ip with 2 × 103 blood trypomastigotes of the Y strain. (A) Myd88−/− (CD45.2+)→WT (B6 x B6.SJL F1, CD45.1+CD45.2+) and WT (B6.SJL, CD45.1+)→WT (B6 x B6.SJL F1, CD45.1+CD45.2+) chimeric mice 8 weeks after reconstitution and (B) WT (B6) and Myd88−/− mice. Survival curves are statistically different (p<0.05). All surviving mice in (A) were euthanized on day 25 pi (n = 6 to 9 per group). (C) Representative dot plots showing frequencies of CD45.1+ (WT) and CD45.2+ (Myd88−/−) among gated CD4+ or CD8+ spleen T cells from control non-infected or infected mixed BM chimeras. (D, E) Mean frequencies of data shown in (C). (F) Representative dot plots showing frequencies of IFN-γ+CD45.1+ and IFN-γ+CD45.2+ among gated CD4+ or CD8+ T cells. (G, H) Mean frequencies of data shown in (F). Mice (n = 4 per group) from control or infected mixed BM chimeric mice were Individually analyzed on day 14 pi. Error bars = SEM, *p≤0.05; **p≤0.01 (two-tailed Student t test). Data are representative of 4 independent experiments. Experimental design, frequency of CD45.1+ (B6.SJL, WT) and CD45.2+ (Myd88−/−) gated on CD11chigh spleen cells, as well as the kinetics of IFN-γ+CD4+ T cell response are shown in Figure 1—figure supplement 1.
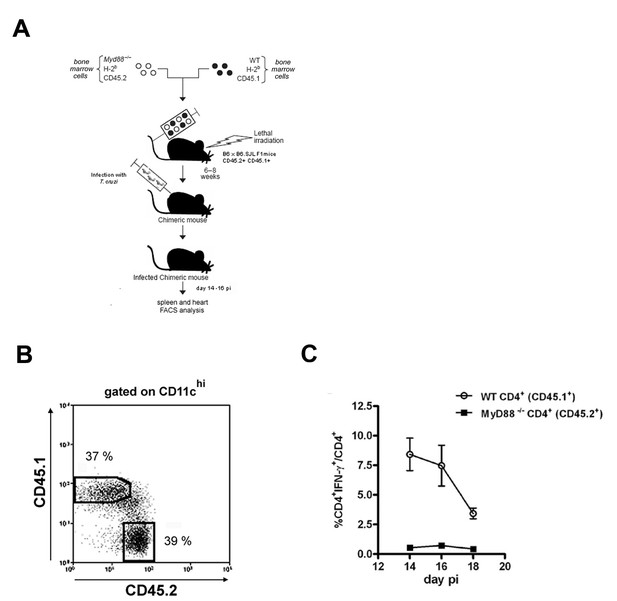
Mixed bone-marrow chimeras.
(A) Experimental design for mixed BM chimera construction. Irradiated WT (B6 x B6.SJL F1) mice were reconstituted with a 1:1 mix of WT (B6.SJL, CD45.1+) and Myd88−/− (CD45.2+) BM cells. (B) Representative frequencies of CD45.1+ (B6.SJL, WT) and CD45.2+ (Myd88−/−) cells gated on CD11chi spleen cells of non-infected mixed BM chimeric mice. (C) Kinetics of the frequency of IFN-γ+CD4+ T cells among CD4+CD45.1+ WT (white circles) and CD4+CD45.2+Myd88−/− (black squares) splenocytes from infected mixed BM chimeric mice.
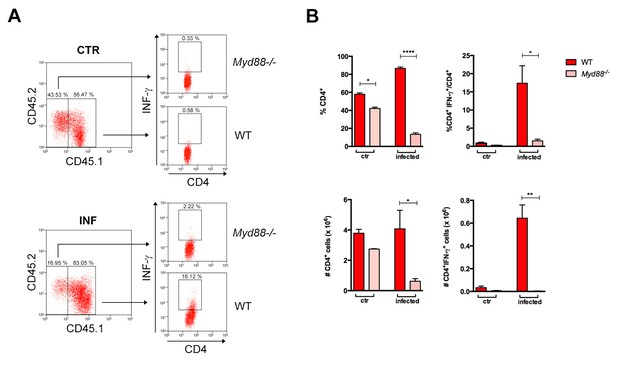
Lower expansion of IFN-γ+CD4+ Myd88 T cells in infected Rag2−/− mice reconstituted with WT and Myd88−/− splenocytes.
(A) Gate strategy and dot plot analysis of IFN-γ+CD4+ T cells among CD4+CD45.2+ (Myd88−/−) or among CD4+CD45.1+ (B6) plus CD4+CD45.2+CD45.1+(B6xB6.SJL F1) (WT) T cells in Rag2−/− mice, reconstituted as described in Material and Methods section. (B) Mean frequencies and absolute cell numbers of Myd88−/− (rose) and WT (red) total CD4+ or IFN-γ+CD4+ T cells gated on CD4+ T cells from reconstituted naïve or infected Rag2−/− mice, individually analyzed on day 14 pi (n = 3 to 4). Error bars = SEM, *p≤0.05; **p≤0.01; ****p≤0.000 (two-tailed Student t-test). Data are representative of 2 independent experiments. Results obtained for the CD8+ T cell subpopulation are shown in Figure 2—figure supplement 1.
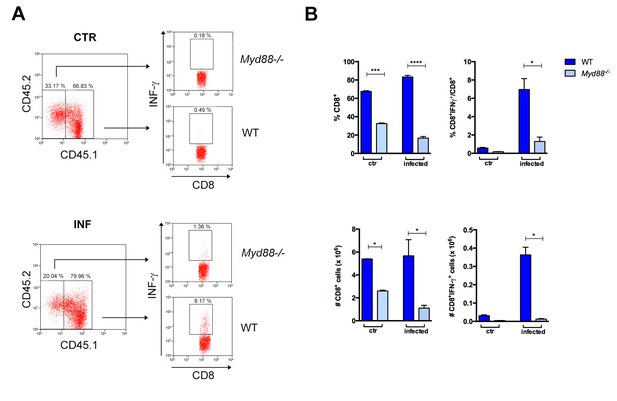
Lower expansion of IFN-γ+CD8+ Myd88-/- cells in infected Rag2−/− mice reconstituted with WT and Myd88−/−splenocytes.
(A) Gate strategy and dot plot analysis of IFN-γ+CD4+ T cells among CD8+CD45.2+ (Myd88−/−) or among CD8+CD45.1+ (B6) plus CD8+CD45.2+CD45.1+(B6xB6.SJL F1) (WT) T cells. (B–C) Mean frequencies and absolute cell numbers of Myd88−/− (light blue) and WT (dark blue) of (B) total CD8+or (C) IFN-γ+CD8+ T cells in reconstituted naïve or infected Rag2−/− mice, individually analyzed on day 14 pi (n = 3 to 4). Error bars = SEM, *p≤0.05; **p≤0.01; ***p≤0.001, ****p≤0.0001 (two-tailed Student t-test). Data are representative of 2 independent experiments.
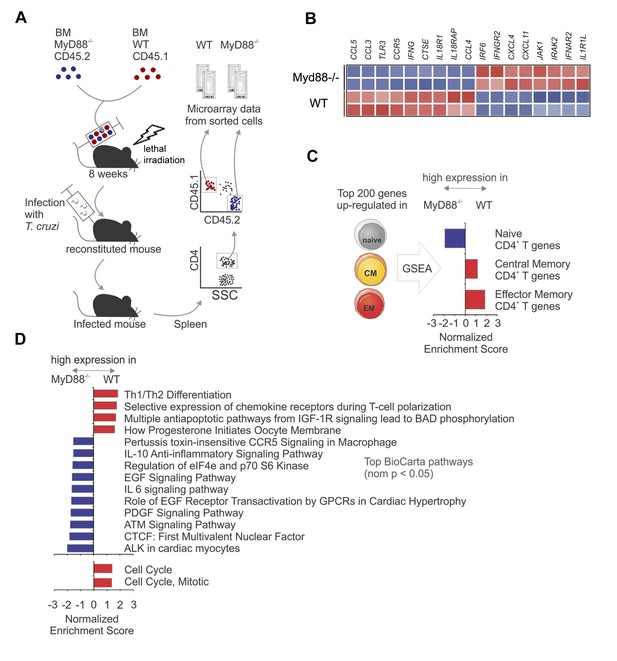
Gene expression profiles of WT and Myd88−/− CD4+T cells sorted from mixed BM chimeras infected with T. cruzi.
(A) Experimental design: CD4+CD45.1+ cells (WT) and CD4+CD45.2+ cells (Myd88−/−) were sorted (99% of purity) from the pool of splenocytes obtained from infected mixed BM chimeric mice at day 14 pi (n = 4). Two individually sorted samples of each cell population (WT or Myd88−/−) were assayed independently for gene expression profile. (B) Heat map of normalized expression of Th1 signature genes in sorted WT or Myd88−/− CD4+ T cells: upregulated in red, downregulated in blue. (C) Gene Set Enrichment Analysis (GSEA) applied to genes highly expressed in WT or Myd88−/− CD4+ T cells using gene signatures of naïve and memory CD4+ T cells. Gene sets from naïve, central memory (CM), effector memory (EM) CD4+ T cells were generated using the top 200 differentially expressed genes in each subset (see Methods). Gene sets enriched in WT cells (red bars) or in Myd88−/− cells (blue bars) are displayed. (D) GSEA using Selected BioCarta (top) and Reactome (bottom) pathways (nominal p-value<0.05) enriched in WT cells (red bars) or in Myd88−/− cells (blue bars) are displayed. GSEA was performed using the pre-ranked genes and parameters as described in (C). Colors represent higher expression in WT (red) or Myd88−/− (blue). Gene networks with genes in core from GSEA analysis are shown in Figure 3—figure supplement 1.
-
Figure 3—source data 1
Table 1. Differential expressed genes between WT and Myd88−/− CD4+ T cells sorted from infected BM-mixed chimeras.
- https://doi.org/10.7554/eLife.30883.008
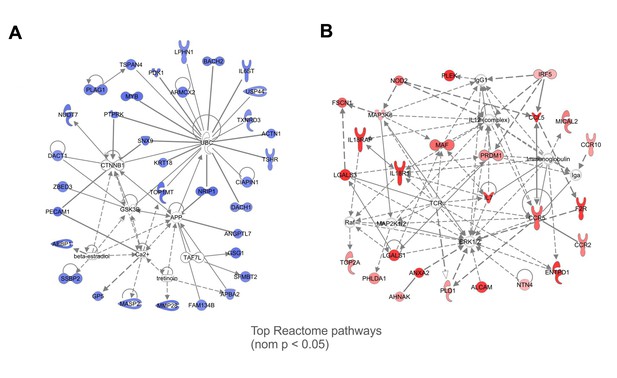
Gene networks with the genes in core from GSEA analysis.
Interactions among the genes were assessed and visualized by Ingenuity Pathway Analysis (IPA) database. Darker colors represent higher expression in (A) Myd88−/− (blue) or in (B) WT (red) CD4+ T cells sorted from the pool of splenocytes obtained from infected mixed BM chimeric mice. Network in (A) represent naïve CD4+ T cell genes highly expressed in Myd88−/− CD4+ T cells. Network in (B) represent EM CD4+ T cell genes highly expressed in WT CD4+ T cells.
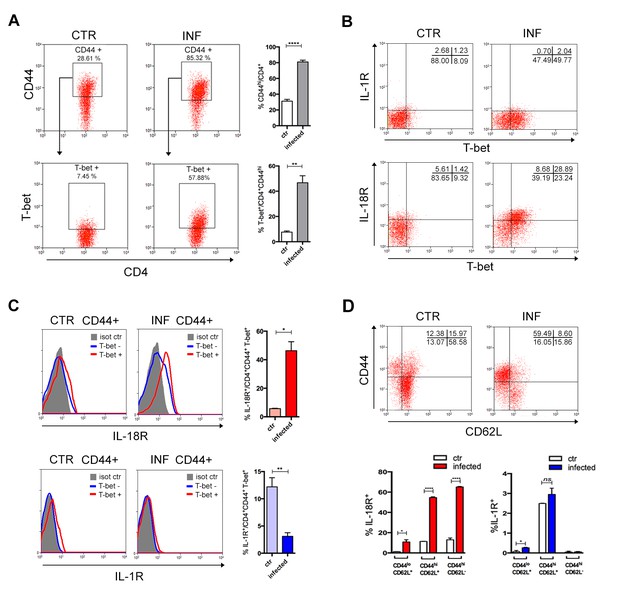
IL-18R, but not IL-1R, is upregulated in CD4+T cells upon activation following infection.
(A) Gate strategy and mean percentage of CD44hiCD4+ and T-bet+CD44hiCD4+ cells gated on CD4+ T splenocytes from non-infected controls and infected B6 mice at day 14 pi. (B) Representative dot plot of IL-1R, IL-18R and T-bet expression, gated on CD44hiCD4+ T cells as in (A). (C) Representative histograms and mean percentages of IL-1R and IL-18R expression, gated on T-bet+CD44hiCD4+ T cells, as shown in (A) and (B). (D) Representative dot plot of CD44 and CD62L expression, gated on CD4+ T cells and mean percentages of IL-1R and IL-18R expression on CD44loCD62L+, CD44hiCD62L- and CD44hiCD62L+ gated on CD4+ T cells (n = 3 to 4). Error bars = SEM, *p≤0.05; **p≤0.01; ****p≤0.0001; ns = non significant (two-tailed Student t-test). Data are representative of 3 independent experiments. Kinetics of IL-1β and IL-18 levels in the serum, as well as of the appearance of IFN-γ+CD4+ T cells in the spleen are shown in Figure 4—figure supplement 1.
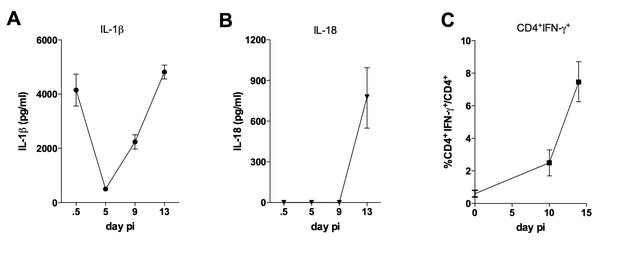
Kinetics of IL-1β and IL-18 levels in the serum, as well as of the appearance of IFN-γ+CD4+T cells in the spleen of infected mice.
(A, B) Serum levels of (A) IL-1β and (B) IL-18. (C) Mean percentages of IFN-γ+CD4+ T cells gated on splenic CD4+ T lymphocytes. Symbols represent the mean value of 4 individually analyzed B6 mice, infected ip with 2 × 103 blood trypomastigotes of the Y strain. Error bars = SEM. Data are representative of 2 independent experiments.
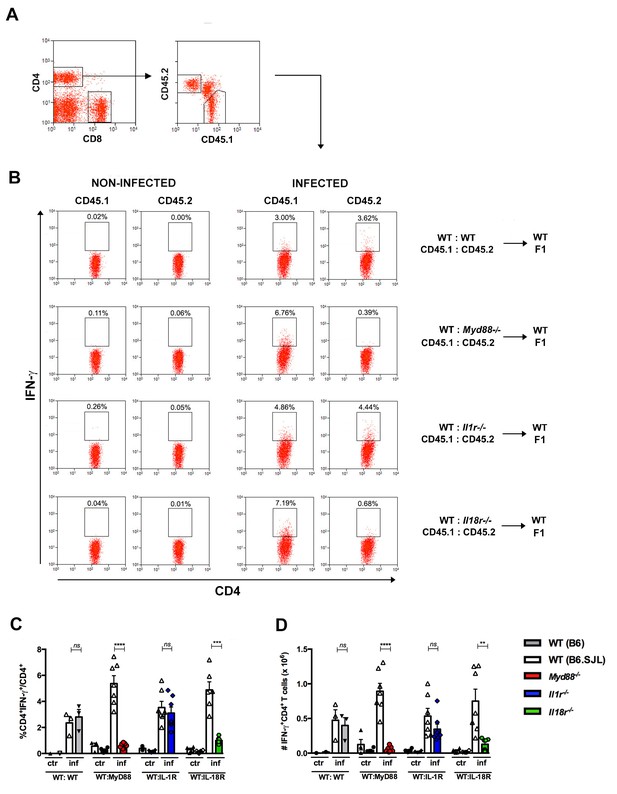
Effects of the absence of T cell-intrinsic MyD88, IL-1R or IL-18R signaling on IFN-γ production by spleen CD4+ T cells of infected mixed BM chimeras.
(A) Representative dot plots showing gating strategy for CD4+ T cells and for CD45.1+ (B6.SJL WT) or CD45.2+ (B6 WT or Myd88−/− or Il1r1−/− or Il18r1−/−) splenocytes from mixed BM chimeras. (B) Representative dot plots of IFN-γ expression gated on CD4+CD45.1+ (B6.SJL WT) or CD4+CD45.2+ (B6 WT or Myd88−/− or Il1r1−/− or Il18r1−/−) splenocytes from non-infected control or infected (2 × 103 blood trypomastigotes, ip) mixed BM chimeras, on day 14 pi. (C) Percentages and (D) absolute cell numbers of IFN-γ+CD4+ T cells gated as in (A) and (B). Bars are the mean of combined data from 2 independent experiments (n = 3 to 7 individually analyzed chimeric mice); error bars = SEM; ns = non significant; **p≤0.01; ***p≤0.001, ****p≤0.0001 (two-tailed Student t-test). Data are representative of 3 to 4 independent experiments.
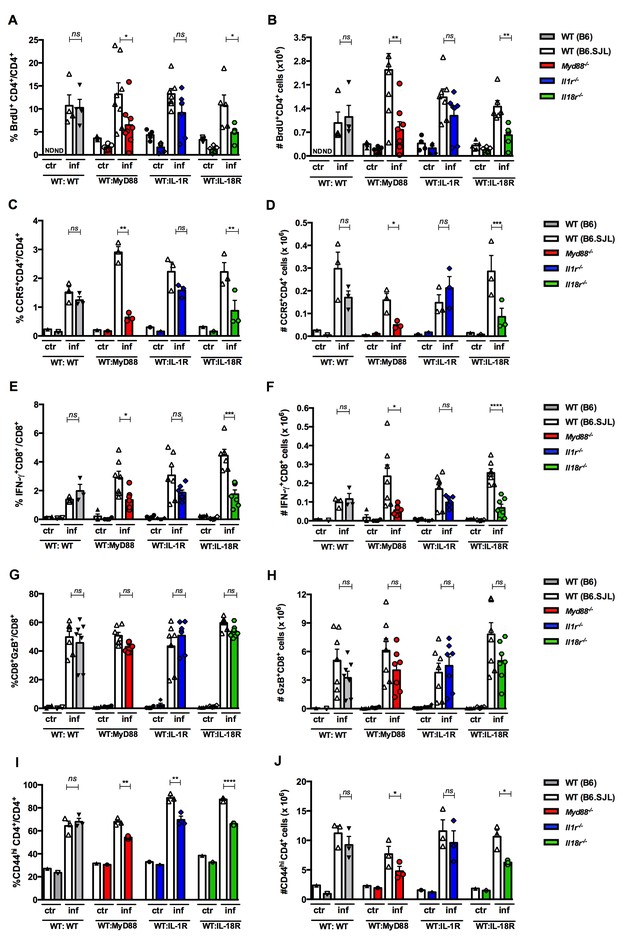
Effects of the absence of T cell-intrinsic MyD88, IL-1R or IL-18R signaling on the proliferation and phenotype of CD4+ and CD8+spleen T cells from infected mixed BM chimeras.
(A, C, G, E and I) Mean frequencies and (B, D, F, H and J) absolute cell numbers of: (A and B) BrdU+CD4+ and (C and D) CCR5+CD4+ cells gated on WT CD45.1+ or KO CD45.2+ CD4+ T cells, as in Figure 5A. (E and F) IFN-γ+CD8+ and (G and H) GzB+CD8+ T cells gated on WT CD45.1+ or KO CD45.2+ CD8+ T cells, as shown in Figure 6—figure supplement 3. (I and J) CD44hiCD4+ T cells, gated on WT or KO CD4+ T cells as in Figure 5A. Bars are the mean of individually analyzed mice from one (C, D, I and J) or two combined independent experiments (A, B, E, F, G and H). (n = 3 to 8); error bars = SEM; ND = not done; ns = non significant; *p≤0.05; **p≤0.01; ***p≤0.001, ****p≤0.0001 (two-tailed Student t-test). Gate strategies and representative dot plots are shown in Figure 6—figure supplements 1–5.
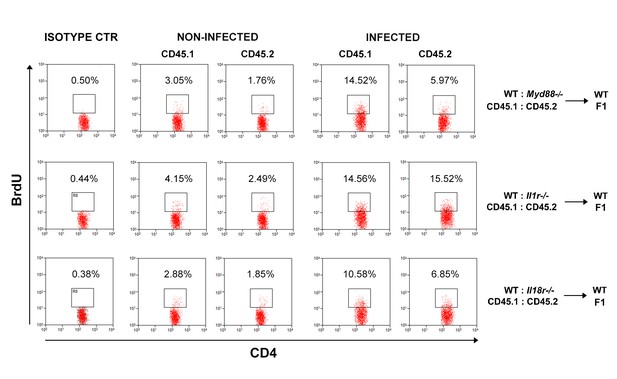
Representative dot plots and gate strategy of BrdU+CD4+T cell analysis from chimeric mice.
Representative dot plots showing anti-BrdU staining in CD4+CD45.1+(B6.SJL, WT) or CD4+CD45.2+ (B6 WT or Myd88−/− or Il1r1−/−or Il18r1−/−) splenocytes from control or infected mixed BM chimeras, gated as in Figure 5A. Data are representative of 3 independent experiments.
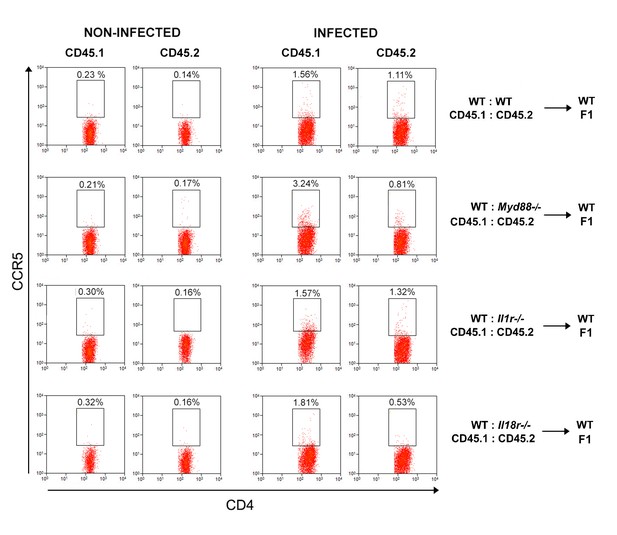
Representative dot plots and gate strategy of CCR5+CD4+ T cell analysis from chimeric mice.
Representative dot plots showing CCR5 expression on CD4+CD45.1+ (B6.SJL, WT) or CD4+CD45.2+ (B6 WT or Myd88−/− or Il1r1−/−or Il18r1−/−) splenocytes, from control or infected mixed BM chimeras, gated as in Figure 5A.
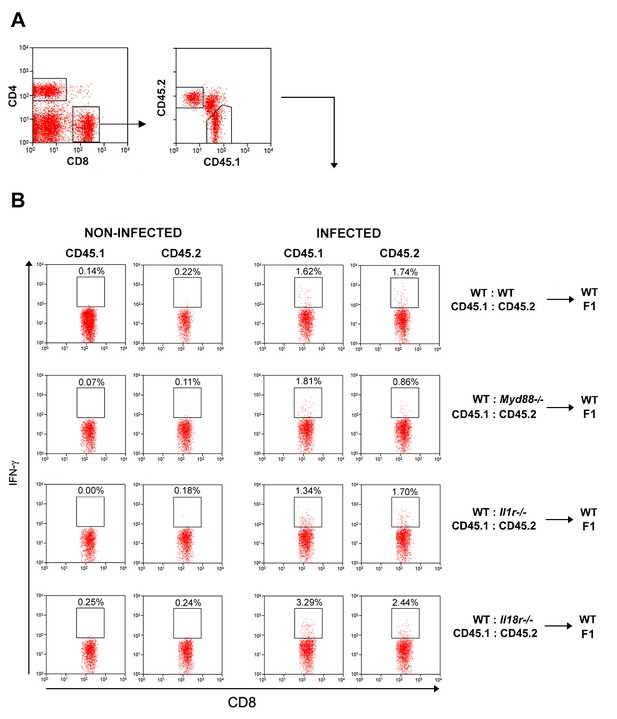
Representative dot plots and gate strategy of IFN-γ+CD8+ T cell analysis from chimeric mice.
(A) Gate strategy for CD45.1+ (B6.SJL WT) and CD45.2+ (B6 WT or Myd88−/− or Il1r1−/−or Il18r1−/−) expression on CD8+ T cells. (B) Representative dot plots of IFN-γ expression gated on CD8+CD45.1+ (B6.SJL WT) or CD8+CD45.2+ (B6 WT or Myd88−/− or Il1r1−/− or Il18r1−/−) splenocytes from non-infected control or infected mixed BM chimeras, gated as in (A). Data are representative of 3 to 4 independent experiments.
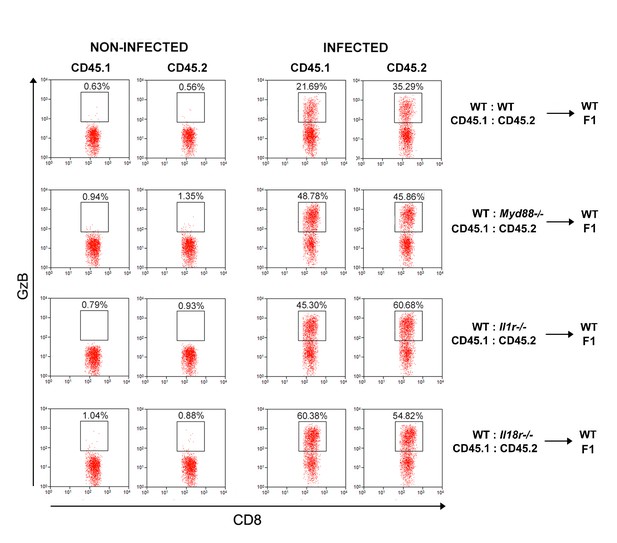
Representative dot plots and gate strategy of GzB+CD8+ T cell analysis from chimeric mice.
Representative dot plots showing anti-GzB staining in CD8+CD45.1+(B6.SJL, WT) or CD8+CD45.2+(B6 WT or Myd88−/− or Il1r1−/− or Il18r1−/−) splenocytes from control or infected mixed BM chimeras, gated as in Figure 6—figure supplement 3. Data are representative of 3 to 4 independent experiments.
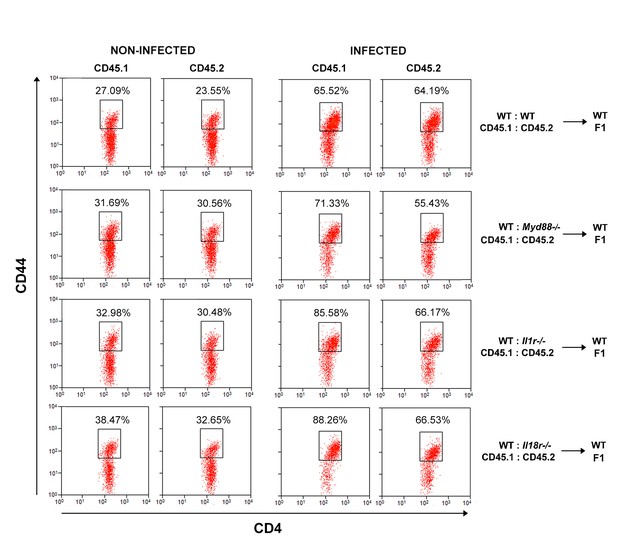
Representative dot plots and gate strategy of CD44hiCD4+ T cell analysis from chimeric mice.
Representative dot plots showing anti-CD44 staining on CD4+CD45.1+(B6.SJL, WT) or CD4+CD45.2+(B6 WT or Myd88−/− or Il1r1−/− or Il18r1−/−) splenocytes from control or infected mixed BM chimeras, gated as in Figure 5A.
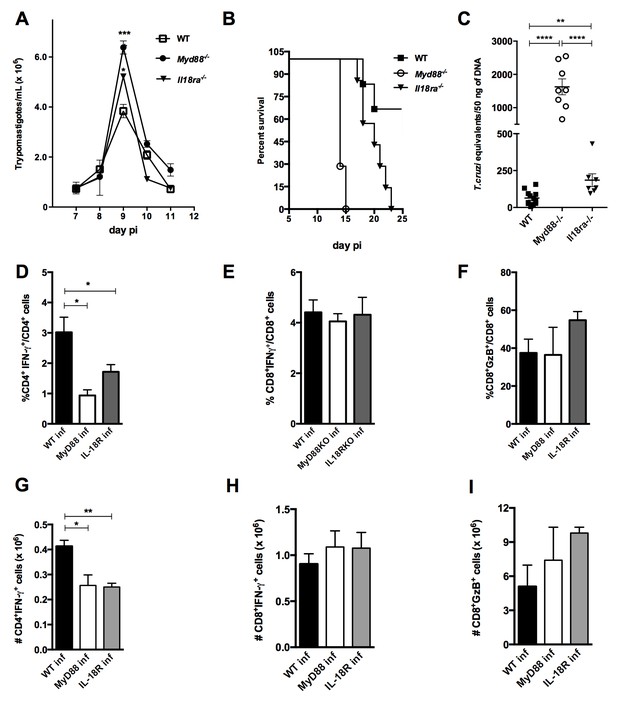
Il18r1−/− mice are highly susceptible to infection with T. cruzi and display lower frequency and absolute numbers of Th1 cells.
(A) Parasitemia curve, (B) survival and (C) parasite load in the myocardium at day 14 pi of Il18r1−/−, Myd88−/− and WT B6 mice infected with 2 × 103 blood trypomastigotes of the Y strain. Survival curves are statistically different (p<0.05). (D–I) Mean frequencies and absolute numbers of (D, G) IFN-γ+CD4+(E, H) IFN-γ+CD8+ and (F, I) GzB+CD8+ cells, gated on CD4+ or CD8+ T splenocytes from 4 to 8 individually analyzed mice at day 14 pi. Data are representative of 3 independent experiments. Error bars = SEM; ns = non significant; *p≤0.05; **p≤0.01; ****p≤0.0001 (two-tailed Student t-test).
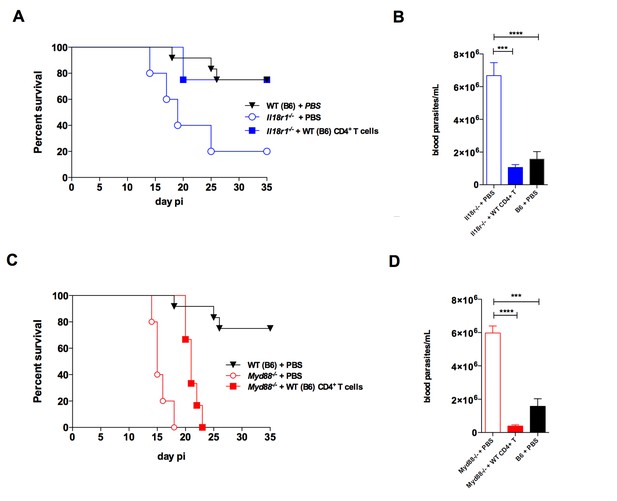
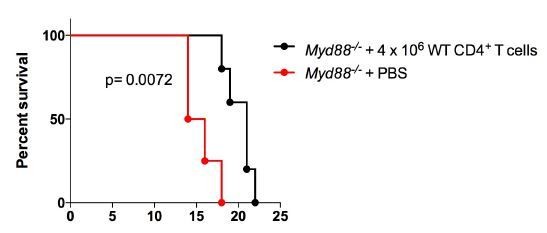
The adoptive transfer of purified WT CD4+ T cells protects Il18r1−/− and Myd88−/− mice against infection with T. cruzi.
(A and C) Survival; (B and D) mean parasitemia levels at day 9 pi of Il18r1−/− (A and B) or Myd88−/− mice (C and D), which received or not 1.3 × 106 CD4+ T cells purified from infected WT B6 mice. Results obtained in infected but non-transferred WT B6 mice are shown in black lines and black bars. Data are representative of 2 independent experiments, using 8 to 10 male mice in each group; error bars = SEM. Survival curves of transferred and non-transferred mice are statistically different (p<0.05). Purity of adoptive transferred CD4+ T cells and parasitemia in Myd88−/− mice adoptively transferred with WT or Ifng−/− CD4+ T cells are shown in Figure 8—figure supplement 1.
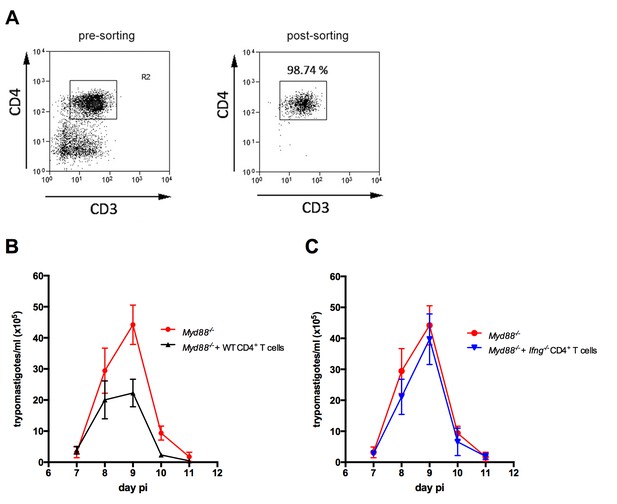
Adoptive transfer of WT, but not of Ifng−/−CD4+ T cells diminished parasitemia in infected Myd88−/− mice.
(A) Sorting of CD4+ T cells: Representative frequency of CD4+CD3+ T cells after enrichment by negative selection with magnetic beads (pre-sorting, left) followed by FACS sorting (post-sorting, right), as described in Materials and Methods. Post-sorted cells (>98% pure) were employed in the adoptive transfer experiments shown in Figure 8. (B–C) Mean parasitemia in Myd88−/− mice, which received or not 1.0 × 106 CD4+ T cells (sorted as in A), purified from infected (B) WT (B6) or (C) Ifng−/− mice. Data are representative of 2 independent experiments; error bars = SEM; (n = 5 male mice individually analyzed).
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30883.021