Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals
Figures
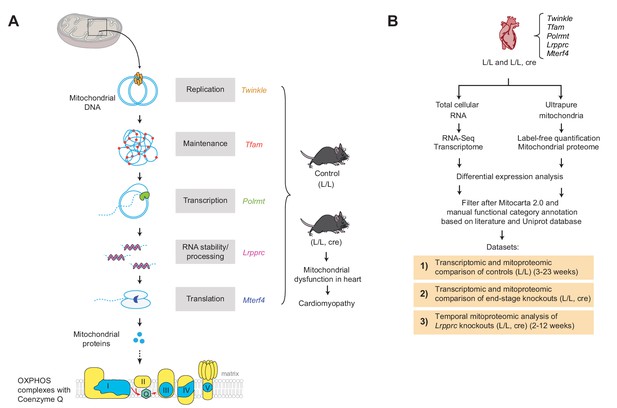
Label-free quantification and comparison of mitoproteomes of mouse hearts with mitochondrial dysfunction.
(A) Schematic representation of the tissue-specific knockout strains (L/L, cre) used, with impaired mtDNA gene expression leading to mitochondrial dysfunction and corresponding controls (L/L). Nuclear-encoded mitochondrial proteins are illustrated in yellow and mtDNA-encoded mitochondrial proteins in blue. (B) Experimental workflow of the analysis of the transcriptomes and mitoproteomes from mouse heart generating three distinct datasets (1-3). Files are provided in Supplementary file 1–5, 8 and 9.
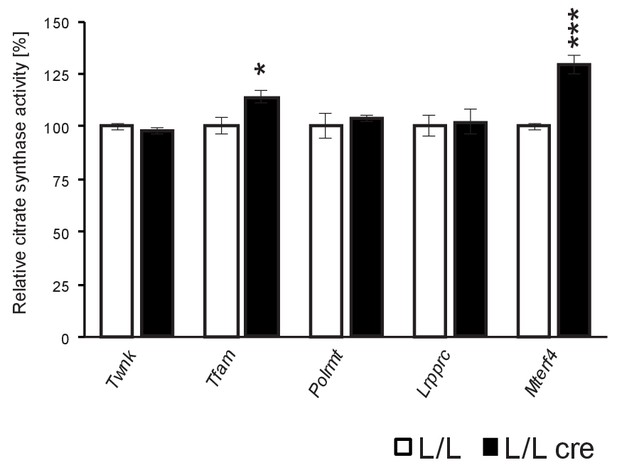
Activity of citrate synthase in heart from different L/L, cre and L/L mice.
Error bars ± SEM; *p<0.05, ***p<0.001; two-tailed unpaired Student’s t-test.
-
Figure 1—figure supplement 1—source data 1
Determination of citrate synthase activity.
- https://doi.org/10.7554/eLife.30952.007
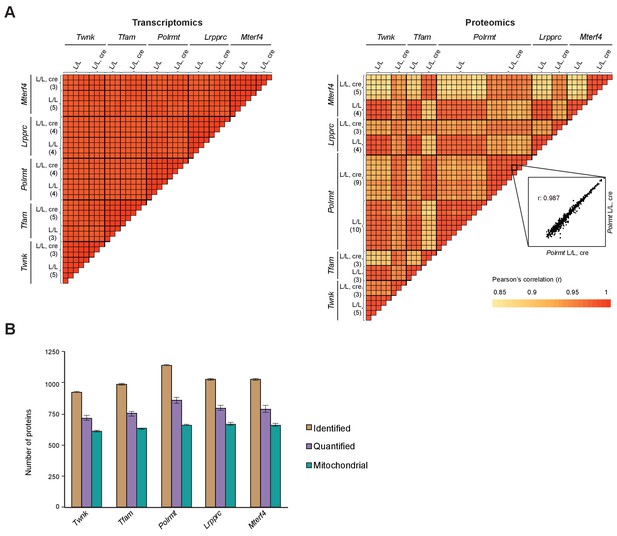
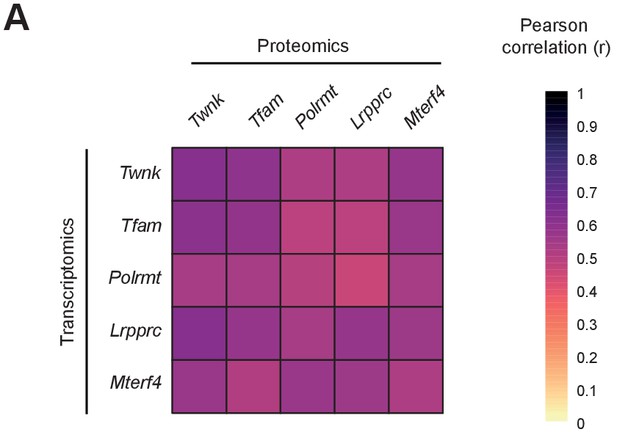
High reproducibility of the transcriptomic and mitoproteomic data.
(A) Reproducibility of the RNA-Seq (left) and label-free mass spectrometry (right) data is illustrated in heatmaps representing the Pearson’s correlation coefficient of all samples of the five different conditional knockouts (L/L, cre) and their corresponding controls (L/L) with the same scale; as an example of reproducibility a scatter plot of the label-free protein intensity values in two Polrmt L/L, cre samples and their correlation coefficient is shown. The number of biological replicates is indicated in parenthesis. (B) Number of identified (851 to 1348 proteins), quantified (673 to 1039 proteins) and mitochondrial proteins (586 to 704 proteins) across all L/L, cre hearts compared to corresponding L/L; error bars: ±SEM.
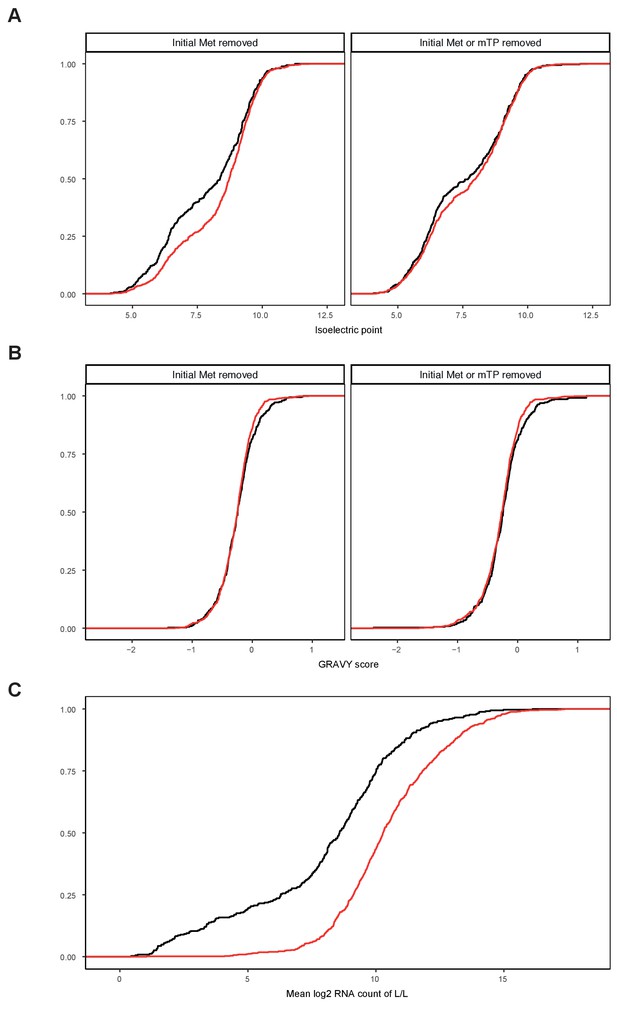
Analysis of systematic bias in the detection of mitochondrial proteins.
Cumulative distribution plots for mitochondrial proteins based on (A) GRAVY score as a measurement of hydrophobicity removing either only the first methionine of the amino acid sequence (left) of the first methionine and the mitochondrial targeting peptide (right). (B) Isoelectric point as a measure of charge hydrophobicity removing either only the first methionine of the amino acid sequence (left) of the first methionine and the mitochondrial targeting peptide (right). (C) mean RNA counts of L/L samples across all mouse strains as a measure of protein abundance. Met, methionine; mTP, mitochondrial targeting peptide; black, detected in at least one knockout mouse strain; red, not detected.
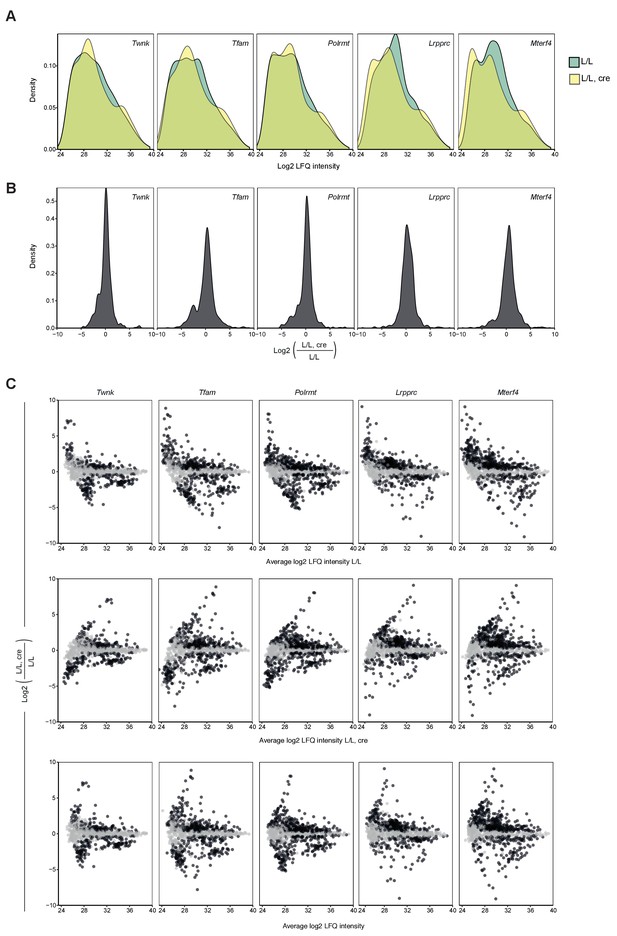
Distribution of label-free quantification (LFQ) intensities and fold changes of quantified proteins.
(A) Distribution of LFQ intensities of quantified proteins on L/L, cre and L/L mitoproteomes per knockout mouse strain. (B) Distribution of fold changes in L/L, cre and L/L mitoproteomes per knockout mouse strain. (C) MA-plots showing the distribution of fold changes in the mitoproteome mice compared to the average LFQ intensity of the proteins in L/L (top), L/L, cre (middle), or both genotypes (bottom) per knockout mouse strain. Black, p<0.05; gray p>0.05.
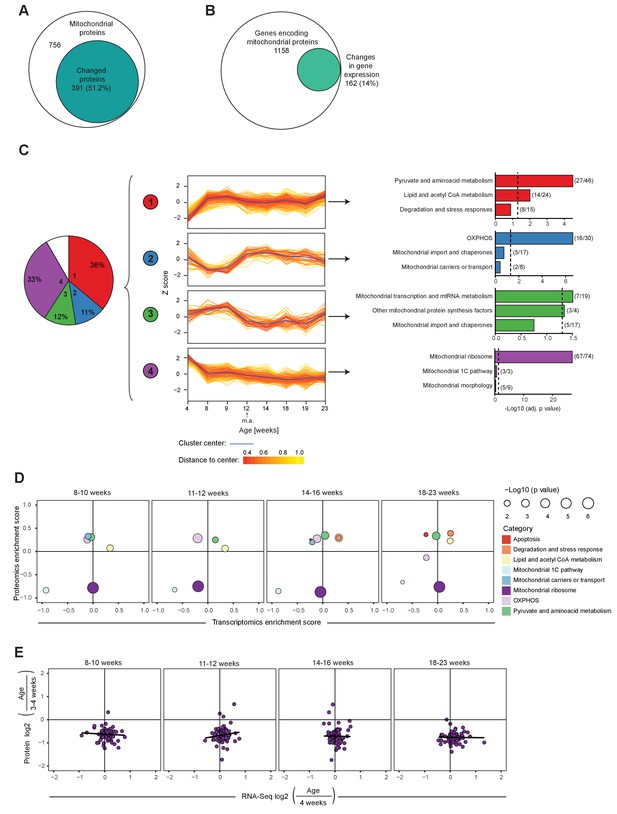
Mitochondrial transcriptome and proteome during post-natal development of mouse heart.
(A–B) Venn diagram of significantly changed (A) mitochondrial proteins and (B) genes encoding for transcripts of mitochondrial proteins of L/L mice (3–20 weeks). (C) Hierarchical clustering analysis of mitoproteomes of L/L mice (3–20 weeks). Left to right: pie chart illustrating percentages of significantly changed mitochondrial proteins in each cluster (1-4), in white: not classified (8%); protein changes over ages for each cluster, fold change relative to 3 week old mice were scaled and presented as Z-score; top three enriched categories of each cluster. Dotted line: Benjamini-Hochberg adjusted p=0.05. Parentheses indicate the number of proteins changed in that category per total number of proteins classified in that category. m.a. = mature adulthood. (D) Mitochondrial transcriptomic and proteomic 2D enrichment analysis showing enriched functional categories of L/L mice at different ages compared to weeks 3–4 based on the fold change. (E) Correlation plots of the L/L, cre versus L/L fold changes between mitoribosomal transcripts and proteins in L/L mice at different ages. Black line indicates the trend.
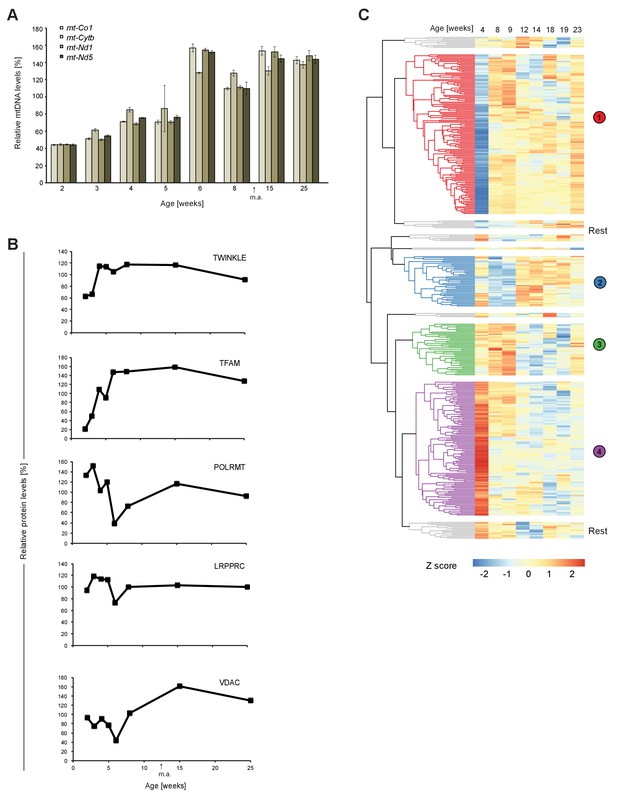
Rapid post-natal increase of mtDNA levels and factors required to maintain and express mtDNA in young wild-type mouse heart.
(A) mtDNA levels in total DNA from wild-type (+/+) mouse heart at different ages. 18S rDNA was used to normalize nuclear input. (B) Immunoblot quantification of some mitochondrial protein levels in heart extracts from wild-type mice at different ages. α-Tubulin was used for normalization. (C) Clustering analysis of significantly changing mitochondrial proteins in control mice (L/L) at different ages. m.a. = mature adulthood.
-
Figure 2—figure supplement 1—source data 1
qPCR determination of mtDNA levels in wild type mice.
- https://doi.org/10.7554/eLife.30952.011
-
Figure 2—figure supplement 1—source data 2
Densitometry analyses of western blots on total proteins.
- https://doi.org/10.7554/eLife.30952.012
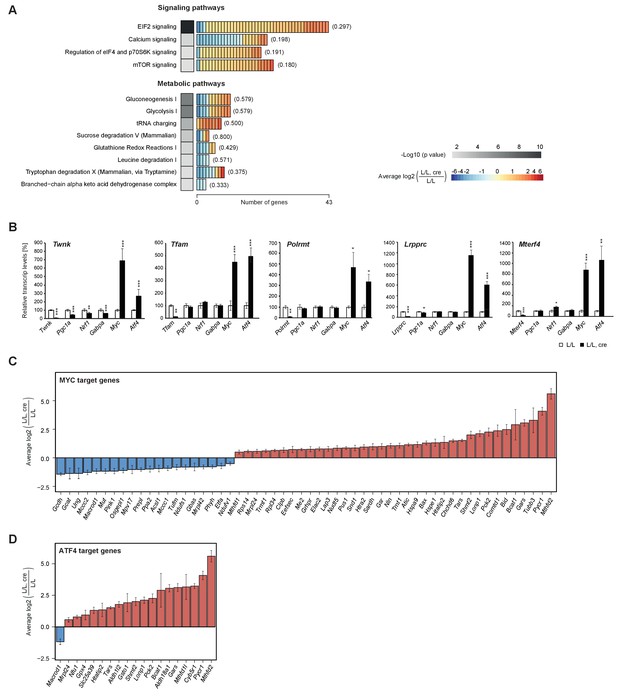
Enrichment of signalling and metabolic pathways in mouse hearts with severe mitochondrial dysfunction.
(A) Canonical pathway analysis of significantly changed genes in all knockouts with representation of the 12 most significant pathways. Grayscale heatmap: p value of each pathway based on Fisher’s exact test. Rectangles in horizontal heatmaps: average expression level in the five knockouts of each gene detected per pathway; Parenthesis: fraction of genes detected per pathway. (B) Transcript levels of genes encoding transcription factors involved in mitochondrial biogenesis in L/L, cre and L/L hearts. Normalization: B2M (beta-2-microglobulin). Twnk, Tfam, Polrmt, Lrpprc, and Mterf4 were used as controls for the corresponding knockout strains. (C–D) Expression levels of differentially expressed MYC and ATF4 target genes encoding mitochondrial proteins. Graphs average expression level in the five knockouts of each gene ± SD.
-
Figure 3—source data 1
qRT-PCR of genes encoding mitochondrial biogenesis factors in the five knockout mouse strains.
- https://doi.org/10.7554/eLife.30952.015
Several targets of MYC and ATF4 transcription factors are differentially regulated upon mitochondrial dysfunction.
Heatmaps illustrate the fold-change transcript levels in L/L, cre and L/L mouse hearts in an alphabetical order. Adjusted p<0.05 in all knockout mouse strains. (A) MYC target genes. (B) ATF4 target genes.
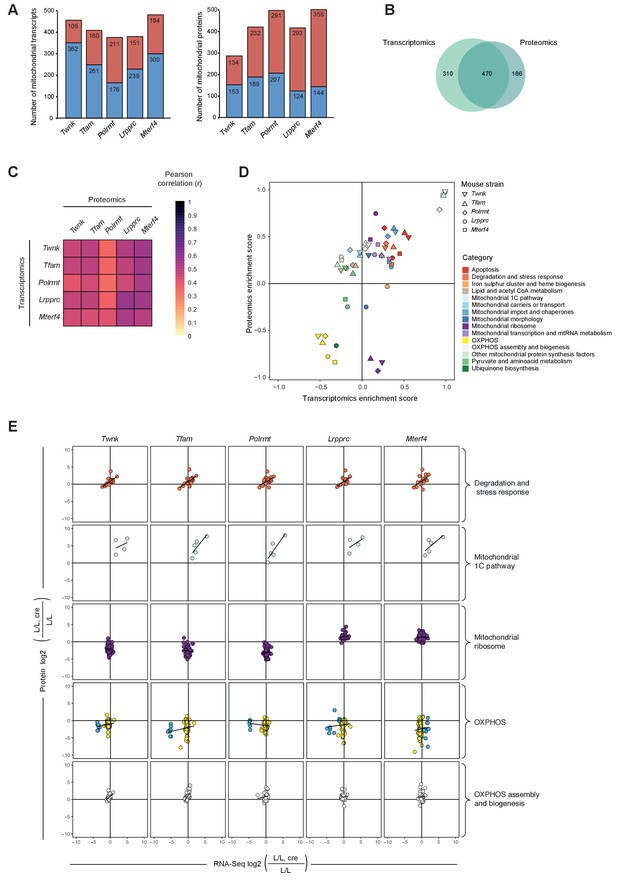
Remodeling of the mitochondrial transcriptome and proteome upon severe mitochondrial dysfunction.
Files are provided in Supplementary files 1 and 4. (A) Number of significantly changed transcripts (left) and mitochondrial proteins (right) quantified from L/L, cre compared to L/L; red: increased, blue: decreased. (B) Venn diagram of number of mitochondrial transcripts and proteins quantified and significant in ≥1 knockout strain. (C) Correlations of the L/L, cre versus L/L fold changes of significantly regulated mitochondrial transcripts and proteins. (D) Mitochondrial transcriptomic and proteomic 2D enrichment analysis showing the trend and degree of regulation of 15 functional categories in all different knockouts. (E) Scatterplots plots of the L/L, cre versus L/L fold changes of mitochondrial transcripts and proteins in different knockouts in a selection of categories. Black line indicates the trend. Same color code applied as in Figure 4D except for the OXPHOS category where the mitochondrial-encoded genes are colored in blue and the nuclear-encoded genes are colored in yellow.
-
Figure 4—source data 1
Pearson correlation coefficient matrixes of fold changes of L/L, cre versus L/L of significantly regulated mitochondrial transcripts and proteins.
- https://doi.org/10.7554/eLife.30952.020
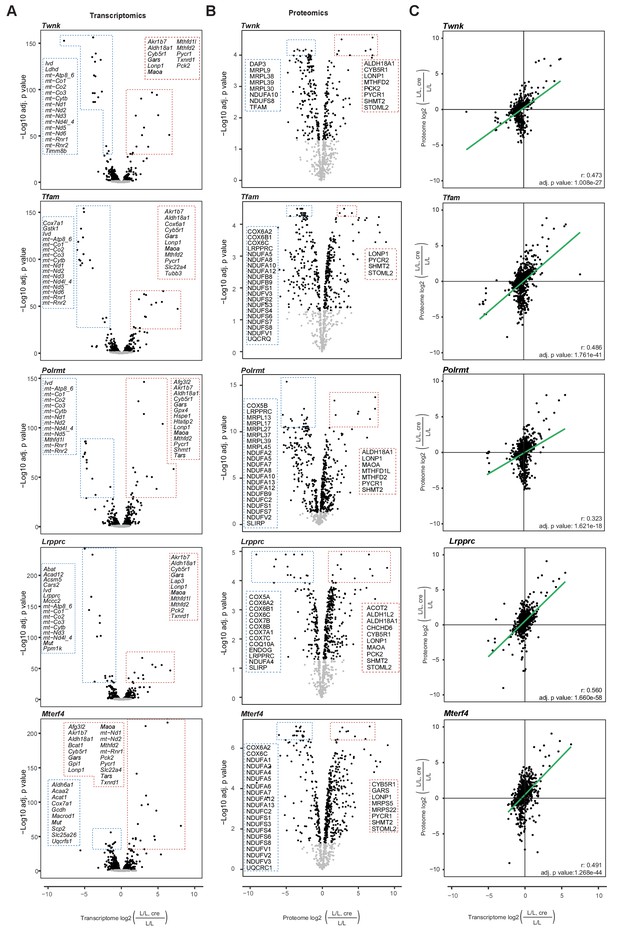
Most of the identified transcripts of nuclear genes encoding mitochondrial proteins are decreased, whereas most of the quantified mitochondrial proteins were increased in abundance.
(A) Volcano plots of transcripts of genes encoding mitochondrial proteins for each knockout mouse strain. (B) Volcano plots of mitoproteomes for each knockout mouse strain. Black: p<0.05, grey: p>0.05. Top 1% of significantly changed mitochondrial transcripts and top 5% of significantly changed mitochondrial proteins are shown and listed in the dotted line boxes; red: increased, blue: decreased; p values were adjusted using Benjamini-Hochberg method. (C) Scatterplots showing the correlation between the mitochondrial transcriptomics and proteomics data for each knockout mouse strain (L/L, cre compared to L/L). Each point corresponds to one quantified gene. Correlation coefficient and p values were calculated with Pearson correlation test.
Correlations of the L/L, cre versus L/L fold changes of significantly regulated mitochondrial transcripts and proteins.
https://doi.org/10.7554/eLife.30952.018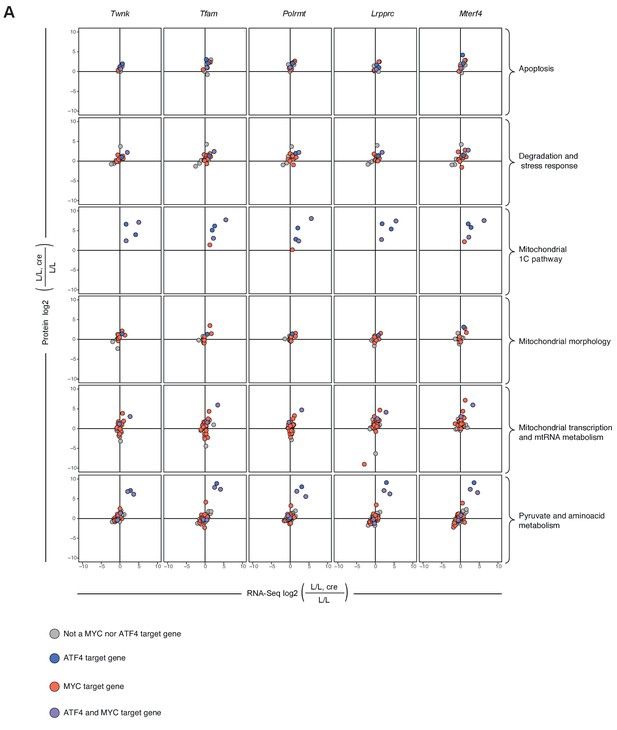
Distribution of MYC and ATF4 target genes in scatterplots plots of the L/L, cre versus L/L fold changes of mitochondrial transcripts and proteins in different knockouts in a selection of categories.
https://doi.org/10.7554/eLife.30952.019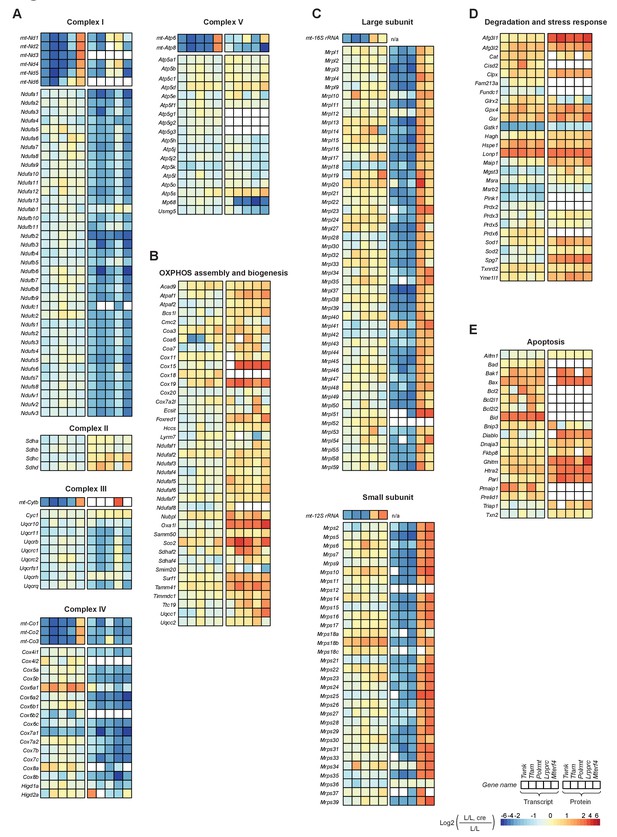
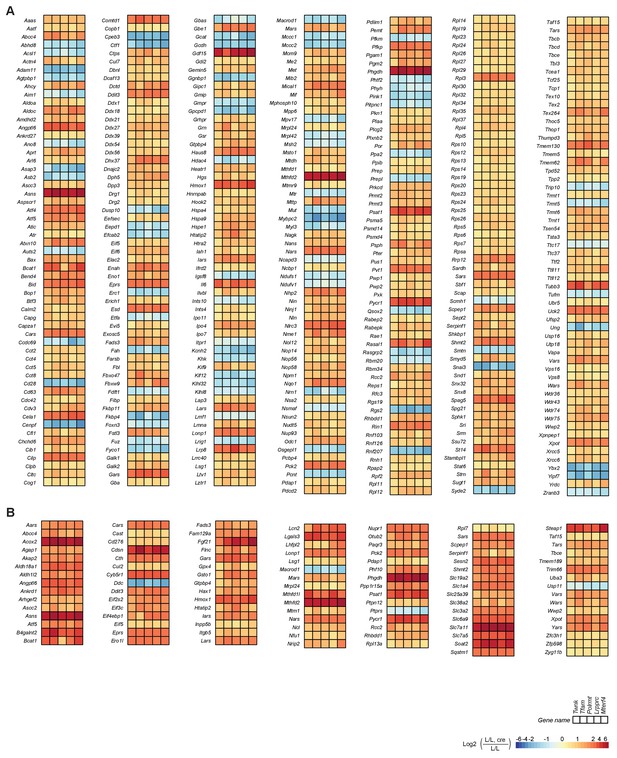
Effects of impaired mtDNA gene expression on mitochondrial protein levels in heart.
Heatmaps illustrating the fold-change transcript (left) and mitochondrial protein (right) levels in L/L, cre and L/L mouse hearts in alphabetical order; blank boxes: not detected or not quantified. Adjusted p<0.05 in≥1 knockout strain. (A) OXPHOS complexes (complex II is only nuclear encoded). (B) OXPHOS assembly. (C) Mitochondrial ribosomal proteins; n/a = not applicable. (D) Degradation and stress response. (E) Apoptosis. For a selection of mitochondrial proteins the steady state levels were verified by immunoblotting shown in Figure 5—figure supplement 2.
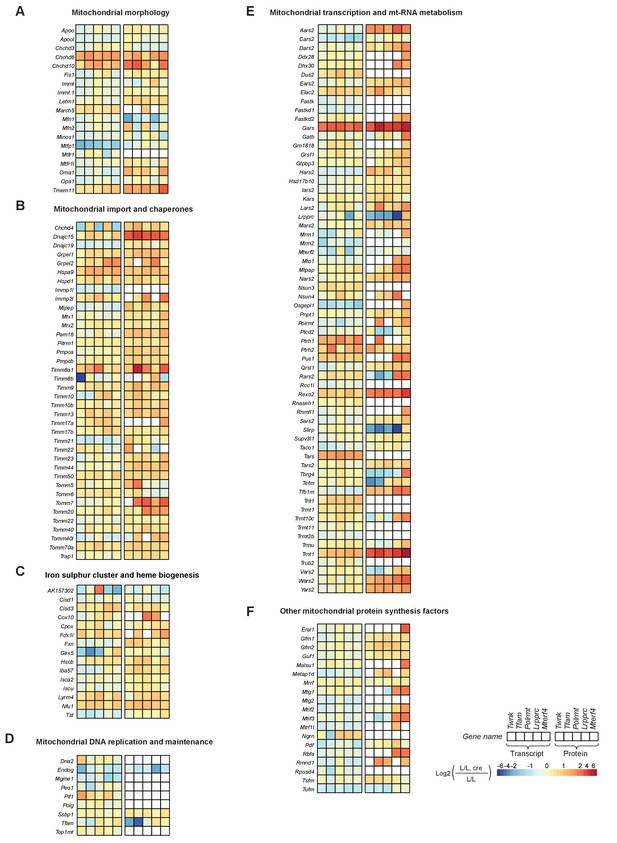
Proteins regulating mitochondrial morphology, iron sulphur cluster and heme biogenesis are increased.
Heatmaps illustrating the fold-change transcript (left) and mitochondrial protein (right) levels in L/L, cre and L/L mouse hearts in alphabetical order; blank boxes in heatmap: not identified or not quantified. Adjusted p<0.05 in at least one of the knockout mouse strains for the genes encoding mitochondrial transcript or protein levels. (A) Mitochondrial morphology. (B) Mitochondrial import and chaperones. (C) Iron sulphur cluster and heme biogenesis. (D) Mitochondrial DNA replication and maintenance. (E) Mitochondrial transcription and mt-RNA metabolism. (F) Other mitochondrial protein synthesis factors.
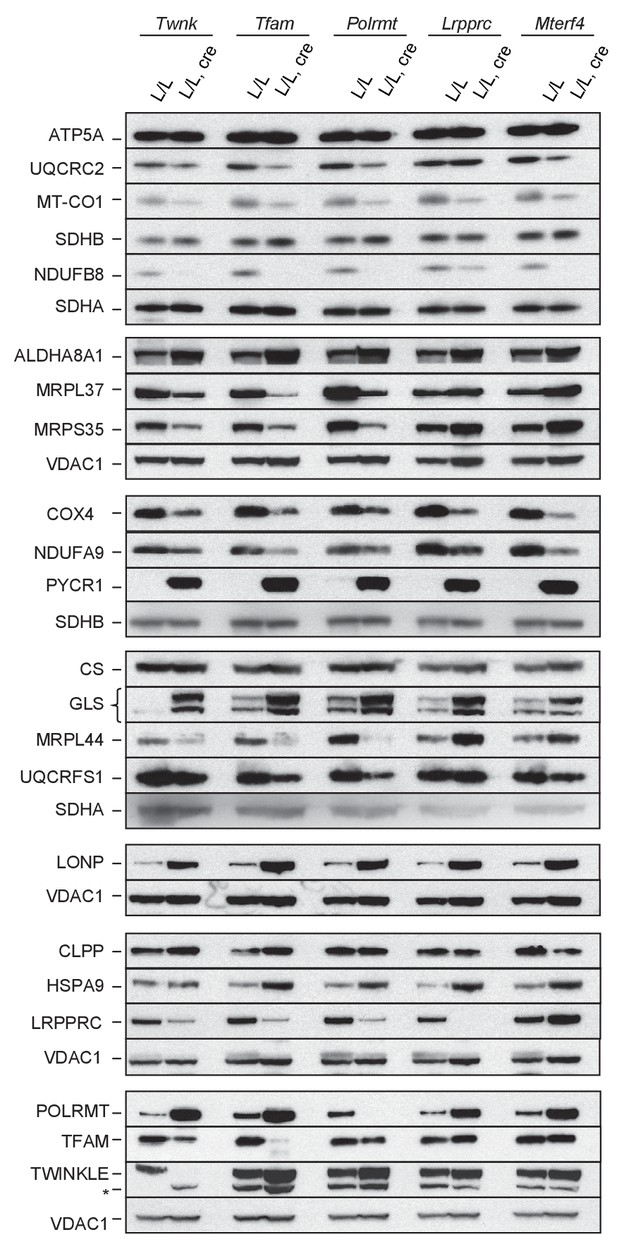
Immunoblot of several mitochondrial proteins in extracts from L/L, cre and L/L hearts; Loading of 6 different membranes: SDHA, SDHB or VDAC1.
https://doi.org/10.7554/eLife.30952.023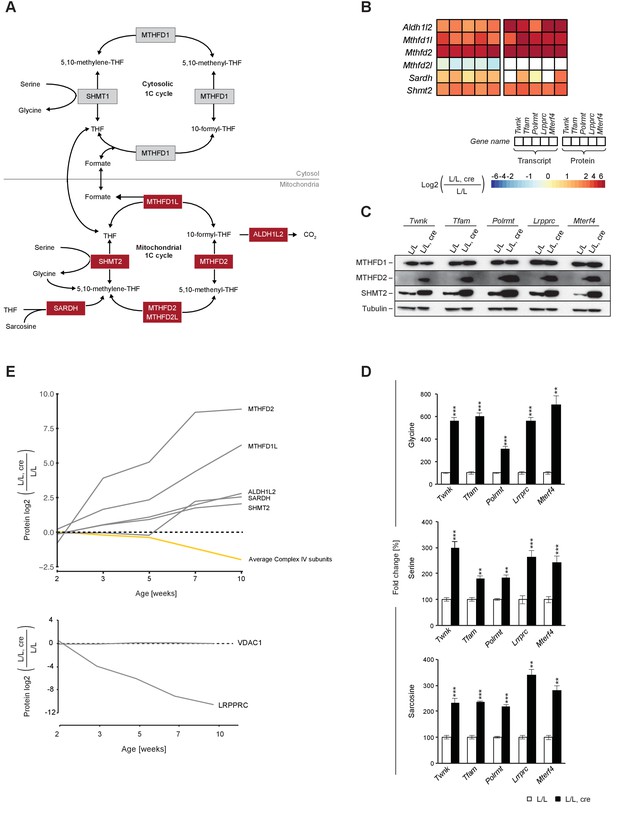
Up-regulation of the enzymes of the mitochondrial 1C pathway happens before deficient OXPHOS is detectable in mouse heart.
(A) Scheme of 1C pathway. Colored boxes: protein levels; red: increased, grey: not detected or not quantified. (B) Heatmaps showing the fold-change in transcript (left) and protein (right) levels in alphabetical order of L/L, cre and L/L mouse hearts of the 1C pathway; p<0.0001 in≥1 knockout strain. (C) Immunoblot of enzymes of the 1C pathway in total protein extracts from L/L, cre and L/L; Loading: tubulin. (D) Quantification of 1C donor metabolite levels in L/L, cre and L/L. Graphs represent mean ± SEM (*p<0.05, **p<0.01, ***p<0.001). (E) Time point analysis of protein levels of enzymes of the 1C pathway (top), and LRPPRC and VDAC (bottom) in Lrpprc knockout hearts compared to controls. Yellow line: average value of nuclear and mitochondrial encoded OXPHOS complex IV subunits. Adjusted p<0.05, except for VDAC. LRPPRC and VDAC protein levels at the different time points were verified by immunoblotting presented in Figure 6—figure supplement 1.
-
Figure 6—source data 1
Determination of 1C pathway donor metabolites.
- https://doi.org/10.7554/eLife.30952.025
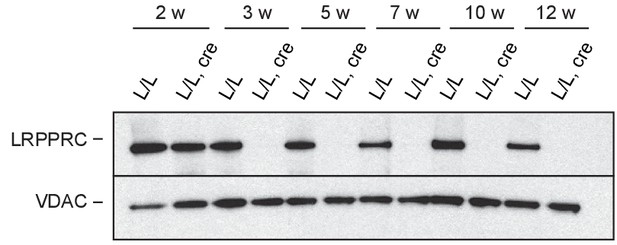
Steady-state LRPPRC protein levels at different time points in mitochondrial extracts from Lrpprc L/L, cre and L/L hearts; Loading: VDAC1.
https://doi.org/10.7554/eLife.30952.026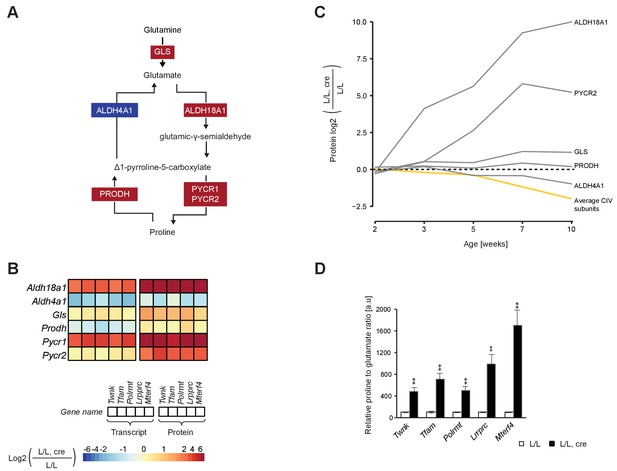
Up-regulated glutamate to proline conversion in mitochondrial OXPHOS deficient heart.
(A) Scheme of the glutamate to proline conversion pathway. Colored boxes: protein levels; red: increased, blue: decreased. (B) Heatmaps illustrating the fold-change transcript (left) and protein (right) levels. Adjusted p<0.05 in≥1 knockout strain for genes encoding mitochondrial transcript or protein levels. (C) Time point analysis of protein levels of enzymes of the glutamate to proline conversion pathway in Lrpprc knockout hearts compared to controls. Yellow line: average value of nuclear and mitochondrial encoded OXPHOS complex IV subunits. Adjusted p<0.05. (D) Quantification of proline and glutamate in different L/L, cre and L/L mouse hearts. Error bars:± SEM; **p<0.01; two-tailed unpaired Student’s t-test.
-
Figure 7—source data 1
Determination of proline and glutamate metabolites.
- https://doi.org/10.7554/eLife.30952.028
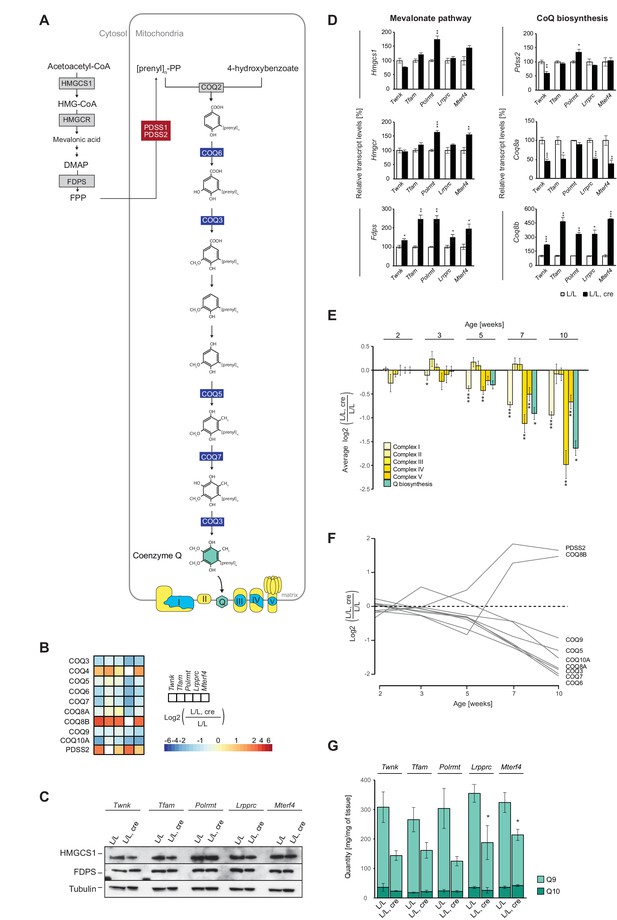
OXPHOS dysfunction leads to decreased cellular Q levels, but the enzymes of the mevalonate pathway are normal.
(A) Scheme of the mevalonate and Q biosynthesis pathways, and OXPHOS complexes (Nuclear-encoded OXPHOS proteins are shown in yellow and mtDNA-encoded OXPHOS proteins in blue). Colored boxes: protein levels; red: increased, blue: decreased, grey: not detected or not quantified. (B) Heatmaps illustrating the fold-change protein levels of the Q biosynthesis pathway in alphabetical order of L/L, cre and L/L mouse hearts; blank boxes: not detected or not quantified proteins; p<0.05 in≥1 knockout strain. (C) Immunoblot of enzymes of the mevalonate pathway on total protein extracts from different L/L, cre and L/L hearts. Loading: tubulin. (D) Transcript levels of genes encoding enzymes of the mevalonate and coenzyme Q synthesis pathway in L/L, cre and L/L hearts. Normalization: B2M (beta-2-microglobulin). (E) Protein levels of OXPHOS complexes I-V and the downregulated Q biosynthesis enzymes at different time points in Lrpprc knockout mouse hearts compared to controls. The graph represents a mean log2 fold-change of all the proteins in that category. (F) Time point analysis of protein levels of enzymes of the Q biosynthesis pathway in Lrpprc knockout mouse hearts compared to controls. Adjusted p across time <0.05. (G) Quinone quantification (Q9 and Q10) in different L/L, cre and L/L mouse hearts. Error bars:± SEM; *p<0.05, **p<0.01, ***p<0.001; two-tailed unpaired Student’s t-test.
-
Figure 8—source data 1
qRT-PCR of genes encoding ubiquinone and mevalonate pathway enzymes in the five knockout mouse strains.
- https://doi.org/10.7554/eLife.30952.031
-
Figure 8—source data 2
Determination of coenzyme Q9 and 10.
- https://doi.org/10.7554/eLife.30952.032
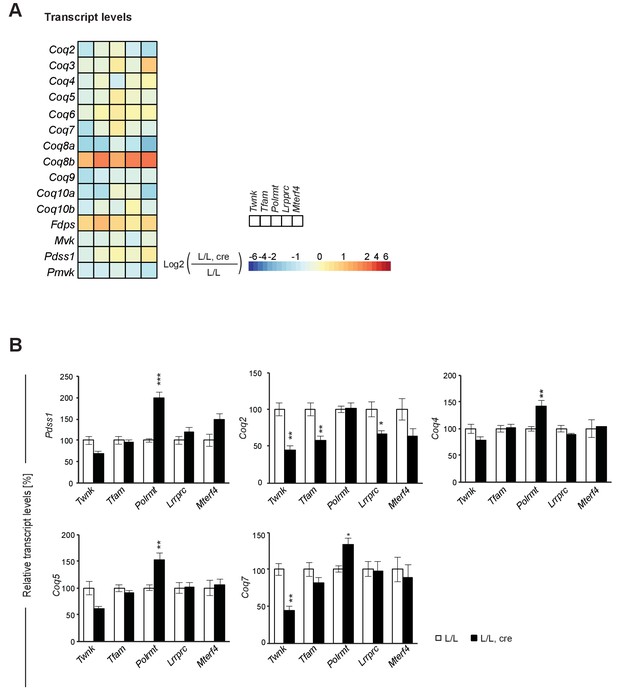
Transcript levels of genes encoding for enzymes of the mevalonate and the Q synthesis pathways.
(A) Heatmaps showing the fold-change transcript levels in alphabetical order of L/L, cre and L/L mouse hearts of the mevalonate and the Q biosynthesis pathways by RNA-Seq.; blank boxes: not detected or not quantified transcript; adjusted p<0.05 in≥1 knockout mouse line. (B) qRT-PCR of transcript levels of genes encoding enzymes of coenzyme Q synthesis pathway in different L/L, cre and L/L hearts, normalized to B2M (beta-2-microglobulin). All graphs represent mean ± SEM; *p<0.05, **p<0.01, ***p<0.001.
Tables
Summary of Major Characteristics of the Five Different Tissue-Specific Knockout Mouse Strains.
Arrows: increase or decrease; tilde: stable. Conditional knockouts: TwnkloxP/loxP, +/Ckmm-cre (Milenkovic et al., 2013), TfamloxP/loxP, +/Ckmm-cre (Larsson et al., 1998), PolrmtloxP/loxP, +/Ckmm-cre (Kühl et al., 2014; Kühl et al., 2016), LrpprcloxP/loxP, +/Ckmm-cre (Ruzzenente et al., 2012), Mterf4loxP/loxP, +/Ckmm-cre (Cámara et al., 2011).
| Conditional knockout (L/L, cre) | Twnk | Tfam | Polrmt | Lrpprc | Mterf4 |
|---|---|---|---|---|---|
| Gene product | Mitochondrial DNA helicase TWINKLE | Mitochondrial transcription factor A | Mitochondrial RNA polymerase | Leucine-rich pentatricopeptide repeat containing protein | Mitochondrial transcription termination factor 4 |
| Lifespan (weeks) | < 19 | < 10 | < 6 | < 16 | < 21 |
| Mitochondrial cardiomyopathy | + | + | + | + | + |
| Mitochondrial DNA | ↓ | ↓ | ↓ | ∼ | ↑ |
| Mitochondrial DNA transcripts | ↓ | ↓ | ↓ | ↓* | ↑ |
| OXPHOS | ↓ | ↓ | ↓ | ↓ | ↓ |
-
* except 12S mt-rRNA, 16S mt-rRNA, mt-Nd6 and most mt-tRNAs
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | ||
|---|---|---|---|---|---|---|
| Organism, C57BL/6N (Mus musculus) | TwnkLoxP/LoxP; Twnk L/L | Milenkovic et al., 2013 | RRID: MGI:5496889 | |||
| Organism, C57BL/6N (Mus musculus) | TwnkLoxP/LoxP, +/Ckmm-cre; Twnk L/L, cre | Milenkovic et al., 2013 | RRID: MGI:5496891 | |||
| Organism, C57BL/6N (Mus musculus) | TfamLoxP/LoxP; Tfam L/L | Larsson et al., 1998 | RRID: MGI:2177633 | |||
| Organism, C57BL/6N (Mus musculus) | TfamLoxP/LoxP, +/Ckmm-cre; Tfam L/L, cre | Larsson et al., 1998 | RRID: MGI:2177634 | |||
| Organism, C57BL/6N (Mus musculus) | PolrmtLoxP/LoxP; Polrmt L/L | Kühl et al., 2014, 2016 | MGI:5704129 | |||
| Organism, C57BL/6N (Mus musculus) | PolrmtLoxP/LoxP, +/Ckmm-cre; Polrmt L/L, cre | Kühl et al., 2014, 2016 | RRID: MGI:5704131 | |||
| Organism, C57BL/6N (Mus musculus) | LrpprcLoxP/LoxP; Lrpprc L/L | Ruzzenente et al., 2012 | RRID: MGI:5438915 | |||
| Organism, C57BL/6N (Mus musculus) | LrpprcLoxP/LoxP, +/Ckmm-cre;Lrpprc L/L, cre | Ruzzenente et al., 2012 | RRID: MGI:5438914 | |||
| Organism, C57BL/6N (Mus musculus) | Mterf4LoxP/LoxP; Mterf4 L/L | Cámara et al., 2011 | RRID: MGI:5288508 | |||
| Organism, C57BL/6N (Mus musculus) | Mterf4LoxP/LoxP, +/Ckmm-cre;Mterf4 L/L, cre | Cámara et al., 2011 | RRID: MGI:5292478 | |||
| Antibody | ALDH18A1 | Thermofisher Scientific | Cat#PA5-19392 RRID: AB_10985670 | (1:200) | ||
| Antibody | CLPP | Sigma-Aldrich | Cat#WH0008192M1 RRID: AB_1840782 | (1:300) | ||
| Antibody | COX4 | Cell Signaling | Cat#4850 RRID: AB_2085424 | (1:500) | ||
| Antibody | CS | Abcam | Cat#ab129095 RRID: AB_11143209 | (1:200) | ||
| Antibody | FDPS | Abcam | Cat#ab189874 RRID: AB_2716301 | (1:500) | ||
| Antibody | GLS | Abcam | Cat#ab93434 RRID: AB_10561964 | (1:200) | ||
| Antibody | HMGCS1 | Abcam | Cat#ab194971 RRID: AB_2716299 | (1:500) | ||
| Antibody | HSPA9/mtHSP70/Grp75 | Abcam | Cat#ab82591 RRID: AB_1860633 | (1:200) | ||
| Antibody | LONP1 | Abcam | Cat#ab103809 RRID: AB_10858161 | (1:500) | ||
| Antibody | LRPPRC mouse | N.-G. Larsson; Ruzzenente et al., 2012 | RRID: AB_2716302 | (1:1000) | ||
| Antibody | MRPL37 | Sigma-Aldrich | Cat#HPA025826 RRID: AB_1854106 | (1:500) | ||
| Antibody | MRLP44 | Proteintech | Cat#16394–1-AP RRID: AB_2146062 | (1:300) | ||
| Antibody | MRPS35 | Proteintech | Cat#16457–1-AP RRID: AB_2146521 | (1:500) | ||
| Antibody | MTHFD1 | Abcam | Cat#ab103698 RRID: AB_10862775 | (1:500) | ||
| Antibody | MTHFD2 | Abcam | Cat#ab37840 RRID: AB_776544 | (1:500) | ||
| Antibody | NDUFA9 | Abcam | Cat#ab14713 RRID: AB_301431 | (1:500) | ||
| Antibody | POLRMT mouse | N.-G. Larsson;Kühl et al., 2014 | RRID: AB_2716297 | |||
| Antibody | PYCR1 | Proteintech | Cat#13108–1-AP RRID: AB_2174878 | (1:200) | ||
| Antibody | SDHA | Thermofisher Scientific | Cat#459200 RRID: AB_2532231 | (1:100) | ||
| Antibody | SHMT2 | Sigma-Aldrich | Cat#HPA020543 RRID: AB_1856833 | (1:500) | ||
| Antibody | TFAM | Abcam | Cat#ab131607 RRID: AB_11154693 | (1:500) | ||
| Antibody | Total OXPHOS Rodent WB Antibody Cocktail | Abcam | Cat#ab110413 RRID: AB_2629281 | (1:1000) | ||
| Antibody | Tubulin | Cell Signaling | Cat#2125 RRID: AB_2619646 | (1:1000) | ||
| Antibody | TWINKLE mouse | N.-G. Larsson,Milenkovic et al., 2013 | RRID: AB_2716298 | |||
| Antibody | UQCRFS1 | Abcam | Cat#ab131152 RRID:AB_2716303 | (1:200) | ||
| Antibody | VDAC1 | Millipore | Cat#MABN504 RRID:AB_2716304 | (1:1000) | ||
| Sequence-based reagent | Taqman Assay - Mouse Adck3 | Life technologies | Mm00469737_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Adck4 | Life technologies | Mm00505363_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Atf4 | Life technologies | Mm00515325_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Myc | Life technologies | Mm00487804_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Coq2 | Life technologies | Mm01203260_g1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Coq4 | Life technologies | Mm00618552_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Coq5 | Life technologies | Mm00518239_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Coq7 | Life technologies | Mm00501587_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Fdps | Life technologies | Mm00836315_g1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Gabpa | Life technologies | Mm00484598_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Hmgcs1 | Life technologies | Mm01304569_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Hmgcr | Life technologies | Mm01282499_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Mterf4 | Life technologies | Mm00508298_m1 | |||
| Sequence-based reagent | Taqman Assay -Mouse Nrf1 | Life technologies | Mm00447996_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Pdss1 | Life technologies | Mm00450958_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Pdss2 | Life technologies | Mm01190168_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Ppargc1 | Life technologies | Mm00447183_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Polrmt | Life technologies | Mm00553272_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Tfam | Life technologies | Mm00627275_m1 | |||
| Sequence-based reagent | Taqman Assay - Mouse Peo1/Twnk | Life technologies | Mm00467928_m1 | |||
| Commercial assay or kit | miRNeasy Mini kit | Qiagen | Cat#217004 | |||
| Commercial assay or kit | Ribo-Zero rRNA removal kit | Illumina | MRZH11124 | |||
| Commercial assay or kit | Tru-Seq Sample preparation | Illumina | RS-122–2002 | |||
| Commercial assay or kit | High Capacity cDNA revese transcription kit | Applied Biosystems | Cat#4368814 | |||
| Commercial assay or kit | TaqMan Universal PCR Master Mix, No Amperase UNG | Applied Biosystems | Cat#4324020 | |||
| Commercial assay or kit | Citrate Synthase Assay Kit | Sigma-Aldrich | Cat#CS0720 | |||
| Chemical compound, drug | EDTA-free complete protease inhibitor cocktail | Roche | Cat#05056489001 | |||
| Chemical compound, drug | PhosSTOP tablets | Roche | Cat#04906837001 | |||
| Chemical compound, drug | Percoll | GE Heathcare | Cat#17-0891-02 | |||
| Chemical compound, drug | Trypsin gold | Promega | Cat#V5280 | |||
| Chemical compound, drug | Standard Coenzyme Q9 | Sigma-Aldrich | Cat#27597 | |||
| Chemical compound, drug | Standard Coenzyme Q10 | Sigma-Aldrich | Cat#C9538 | |||
| Chemical compound, drug | Standard Glutamate (Glutamic acid) | Sigma-Aldrich | Cat#G1251 | |||
| Chemical compound, drug | Standard Glycine | Sigma-Aldrich | Cat#G7126 | |||
| Chemical compound, drug | Standard Proline | Sigma-Aldrich | Cat#P0380 | |||
| Chemical compound, drug | Standard Sarcosine | Sigma-Aldrich | Cat#S7672 | |||
| Chemical compound, drug | Standard Serine | Sigma-Aldrich | Cat#S4500 | |||
| Software, algorithm | Cytoscape v. 3.5.0 | Shannon et al., 2003 | http://www.cytoscape.org RRID:SCR_003032 | |||
| Software, algorithm | DESeq2 package R v. 3.3.2 | Other | Love et al. (2014) | |||
| Software, algorithm | Ingenuity Pathway Analysis - Ingenuity Systems | Qiagen | www.ingenuity.com RRID:SCR_008653 | |||
| Software, algorithm | iRegulon v. 1.3 | Janky et al., 2014 | http://iregulon.aertslab.org/download.html | |||
| Software, algorithm | MaxQuant v. 1.5.2.8 | Cox and Mann, 2008 | http://www.coxdocs.org/doku.php id=maxquant:start RRID:SCR_014485 | |||
| Software, algorithm | R - The R project for Statistical Computing | https://www.r-project.org RRID:SCR_001905 | ||||
| Software, algorithm | Perseus | Cox and Mann, 2012 | http://www.coxdocs.org/doku.php?id=perseus:start | |||
| Software, algorithm | TargetP v. 1.1 | Emanuelsson et al., 2000; Nielsen et al., 1997 | http://www.cbs.dtu.dk/services/TargetP/ | |||
| Other | 1.9 mm ReproSil-Pur 120 C18-AQ media | Dr. Maisch | Cat#r119.aq | |||
| Other | 25 cm, (75 mm internal diameter) PicoFrit analytical column | New Objective | Cat#PF7508250 | |||
Additional files
-
Supplementary file 1
Comparative analysis of mitoproteomic data from heart of Tfam, Twnk, Polrmt, Lrpprc and Mterf4 knockout mice and corresponding controls.
- https://doi.org/10.7554/eLife.30952.033
-
Supplementary file 2
Analysis of mitoproteomic data from heart at different ages of Lrpprc knockout mice and controls.
- https://doi.org/10.7554/eLife.30952.034
-
Supplementary file 3
Analysis of mitoproteomic data from heart at different ages of control mouse strains.
- https://doi.org/10.7554/eLife.30952.035
-
Supplementary file 4
Analysis of total cellular transcriptome from heart of Tfam, Twnk, Polrmt, Lrpprc and Mterf4 knockout and control mouse strains at different ages.
- https://doi.org/10.7554/eLife.30952.036
-
Supplementary file 5
Number of biological replicates and p values of qRT-PCR, metabolomic analyses and enzyme activity measurements.
- https://doi.org/10.7554/eLife.30952.037
-
Supplementary file 6
iRegulon analysis of RNA-Seq data of total RNA from hearts of end-stage conditional knockout mice.
- https://doi.org/10.7554/eLife.30952.038
-
Supplementary file 7
Analysis of proteomic bias in mitoproteomics data from heart of Tfam, Twnk, Polrmt, Lrpprc and Mterf4 knockout mice and corresponding controls.
- https://doi.org/10.7554/eLife.30952.039
-
Supplementary file 8
Complete set of differential expression proteomic analysis in heart of the five knockout mouse strains and according controls; boxplots of the intensity detected by mass spectrometry per protein.
- https://doi.org/10.7554/eLife.30952.040
-
Supplementary file 9
Complete set of sequential mitoproteomic changes at different time points of progressive mitocondrial dysfunction in heart of one knockout mouse strain.
Time curves of differential expression analysis of each protein on the Lrpprc knockout analysis at different ages.
- https://doi.org/10.7554/eLife.30952.041
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30952.042





