Metformin extends C. elegans lifespan through lysosomal pathway
Figures
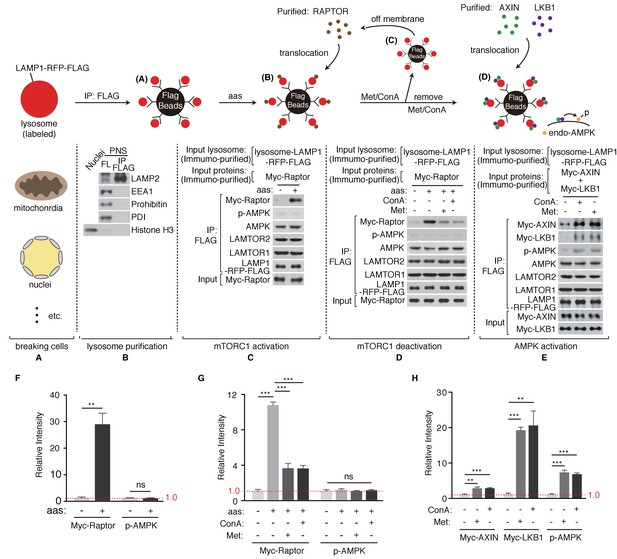
Metformin coordinates mTORC1 and AMPK on purified lysosome.
(A) HEK293T cells stably expressing LAMP1-RFP-FLAG were mechanically broken. (B) Lysosomes were purified through immunoprecipitation (Organelle markers: LAMP2, lysosome; EEA1, early endosome; Prohibitin, mitochondria; PDI, ER; Histone H3, nucleus). (C) Lysosomal accumulation of purified Myc-raptor upon amino acids stimulation. (D–E) Lysosomal disassociation of Myc-raptor (D), lysosomal accumulation of Myc-AXIN/LKB1 and phosphorylation of AMPK (E) upon Concanamycin A (Con A) or Metformin (Met) treatment. (F–H) Quantifications of immunoblots in (C–E). Immunoblots of Myc-Raptor, Myc-AXIN and Myc-LKB1 were normalized to that of LAMP-RFP-FLAG, and immunoblots of p-AMPK were normalized to that of AMPK. Relative intensities of three independent biological replicates are shown as mean ± SEM. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
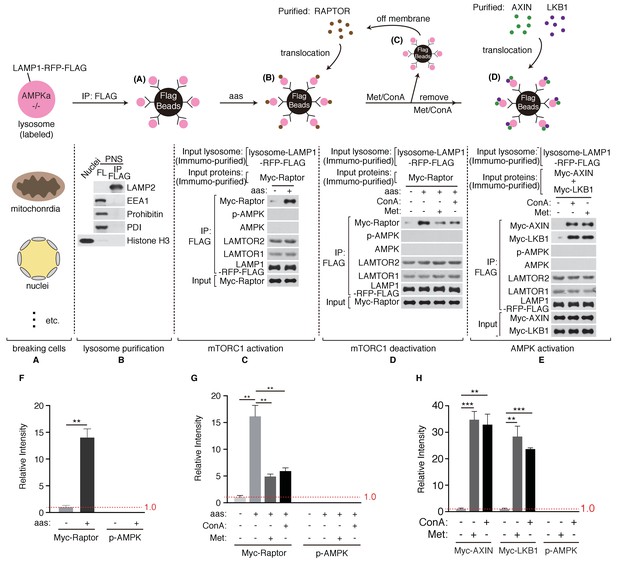
Metformin inhibits mTORC1 independent of AMPK.
(A) AMPKα1/2 double knockout cells stably expressing LAMP1-RFP-FLAG were mechanically broken. (B) Lysosomes were purified through immunoprecipitation (Organelle markers: LAMP2, lysosome; EEA1, early endosome; Prohibitin, mitochondria; PDI, ER; Histone H3, nucleus). (C) Lysosomal accumulation of purified Myc-raptor upon amino acids stimulation. (D–E) Lysosomal disassociation of Myc-raptor (D) and lysosomal accumulation of Myc-AXIN/LKB1 (E) upon Concanamycin A (Con A) or Metformin (Met) treatment. (F–H) Quantifications of immunoblots in (C–E). Immunoblots of Myc-Raptor, Myc-AXIN and Myc-LKB1 were normalized to that of LAMP-RFP-FLAG, and immunoblots of p-AMPK were normalized to that of AMPK. Relative intensities of three independent biological replicates are shown as mean ± SEM. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
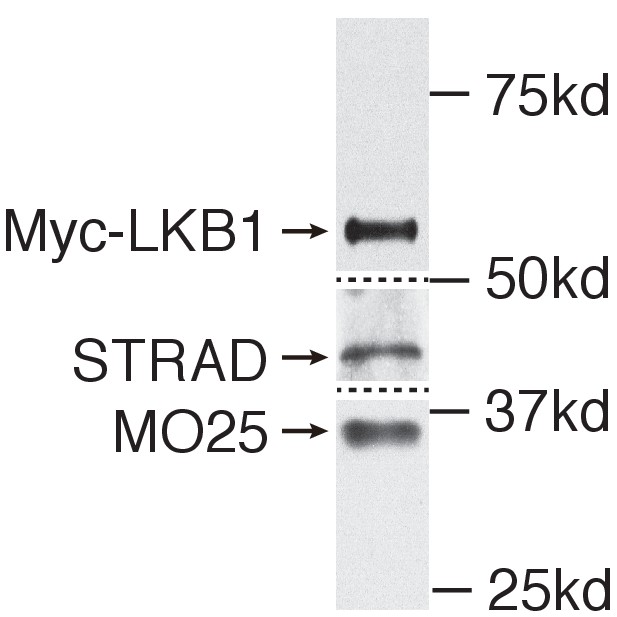
Purified Myc-LKB1 forms a complex with endogenous STRAD and MO25.
https://doi.org/10.7554/eLife.31268.005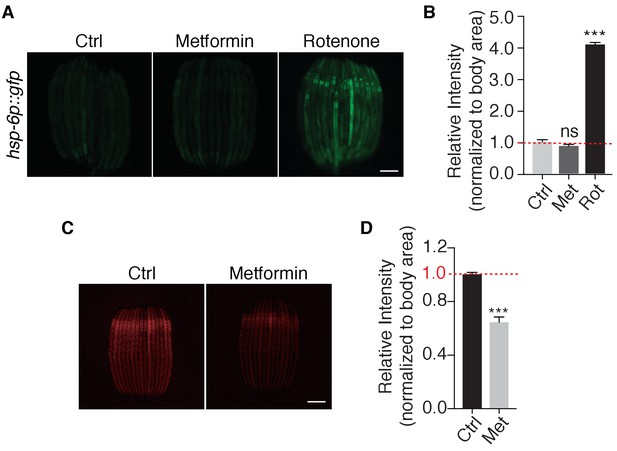
Metformin targets lysosome in C.elegans.
(A) Representative fluorescent images of mitochondrial stress response reporter strain hsp-6p::gfp cultured under normal condition, or in the presence of metformin or rotenone. (B) Quantification of fluorescent images in (A). Mean ± SEM of 3 independent biological replicates are shown (sample size:≥40 worms). (C) Representative fluorescent images of Magic Red Cathepsin assay in worms administrated with or without metformin. (D) Quantification of fluorescent images in (C). Mean ± SEM of 3 independent biological replicates are shown (sample size:≥40 worms). Scale bar: 100 um. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
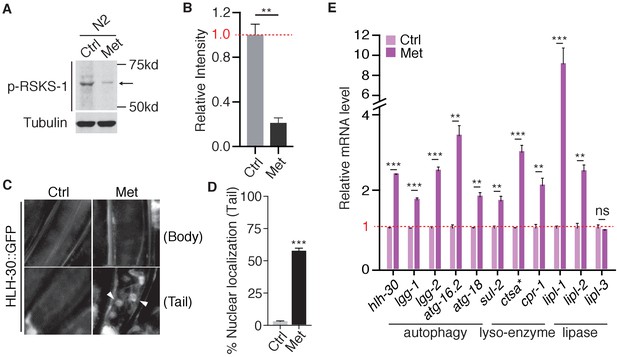
Metformin inhibits TORC1 pathway in C.elegans.
(A) Representative western blotting of RSKS-1 phosphorylation in the presence or absence of Metformin. (B) Quantification of immunoblots in (A). Relative intensities of 3 independent biological replicates are shown as mean ± SEM. (C) Representative fluorescent images of HLH-30 nuclei localization in the presence or absence of metformin. (D) Percentage of worms with HLH-30 nuclear localization in (C) was quantified. Mean ± SEM of 3 independent biological replicates are shown (sample size:≥40 worms). (E) Q-PCR analysis of HLH-30 target genes in the presence or absence of metformin. ~300 worms were pooled in each sample. Data from three independent biological replicates are shown as mean ± SEM. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
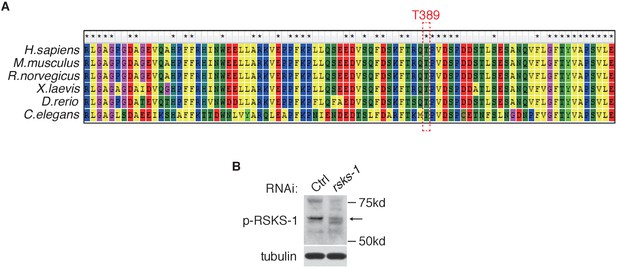
RSKS-1, C.elegans homolog of S6K1 is phosphorylated at T389 residue.
(A) T389 of S6K1 is highly conserved among species. A ClastW of S6K1 protein sequence in different species was plotted by MEGA6. (B) rsks-1 RNAi reduces RSKS-1 phosphorylation level in wild type animals.
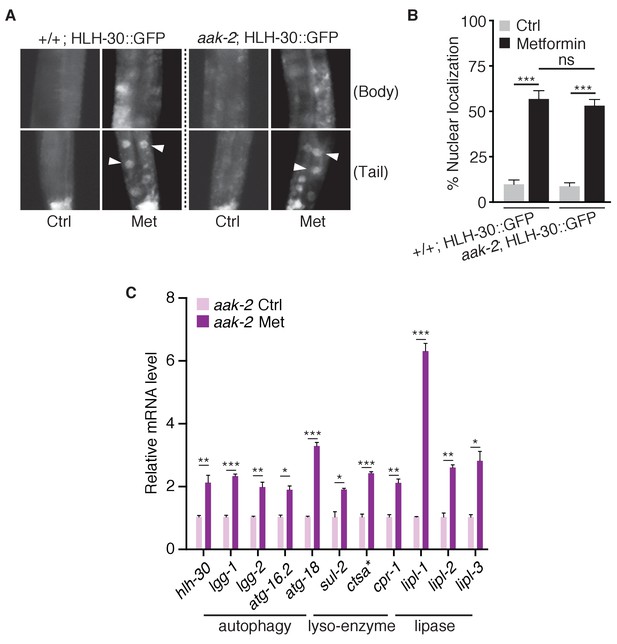
Metformin inhibits TORC1 independent of AMPK in C.elegans.
(A) Representative fluorescent images of HLH-30 nuclei localization in wild type or aak-2 mutant animals in the presence or absence of metformin. (B) Percentage of worms with HLH-30 nuclear localization in (A) was quantified. Mean ± SEM of 3 independent biological replicates are shown (sample size: ≥40 worms). (C) Q-PCR analysis of HLH-30 target genes in aak-2 mutants with or without metformin treatment. ~300 worms were pooled in each sample. Data from three independent biological replicates are shown as mean ± SEM. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
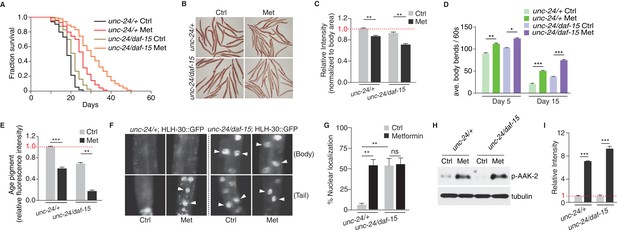
Metformin extends healthspan partially due to TORC1 inhibition.
(A) Lifespan analysis of control worms or daf-15 heterozygous mutants in the presence or absence of metformin. (B) Representative images of Oil-Red-O (ORO) staining of control worms or daf-15 heterozygous mutants in the presence or absence of metformin. (C) Quantification of (B). Error bars represent mean ± SEM of 3 independent biological replicates (sample size: n ≥ 40 worms). (D–E) Locomotion (D) and age pigments (E) were measured in control worms or daf-15 heterozygous mutants in the presence or absence of metformin. Mean ± SEM of 3 independent biological replicates are shown (sample size: n ≥ 20 worms for locomotion assay; n ≥ 40 worms for age pigments assay). (F) Representative fluorescent images of HLH-30 nuclei localization in control worms or daf-15 heterozygous mutants in the presence or absence of metformin. Arrows indicate nuclear localized HLH-30::GFP. (G) Quantification of (F). Percentage of unc-24/+; HLH-30::GFP or unc-24/daf-15; HLH-30::GFP worms with nuclear accumulation of HLH-30 were counted. Error bars represent mean ± SEM of 3 independent biological replicates. (sample size: n ≥ 40 worms) (H) Representative western blotting of AAK-2 phosphorylation in the presence or absence of metformin. (I) Quantification of (H).~300 worms were pooled in each protein sample. Error bars represent mean ± SEM of 3 independent biological replicates. ns, no significant difference; *p<0.05, **p<0.01, ***p<0.001.
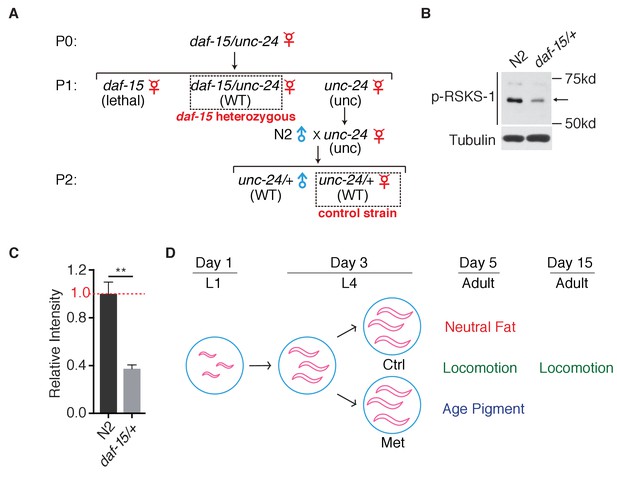
Deletion of daf-15 suppresses TORC1 activity.
(A) Generation of daf-15 heterozygous mutants and its corresponding control animals. (B) Representative immunoblottings of RSKS-1 phosphorylation in N2 or daf-15 heterozygous mutants. (C) Quantification of immunoblots in (B). Relative intensities of 3 independent biological replicates are shown as mean ± SEM. (D) Schematic representation of experimental design for the detection of healthspan parameters. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
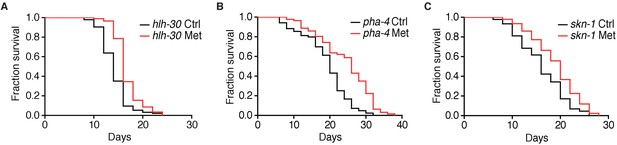
Metformin promotes longevity through a pathway additive to TORC1 inhibition.
(A–C) Lifespan analysis of hlh-30, pha-4 or skn-1 loss-of-function mutants in the presence or absence of metformin.
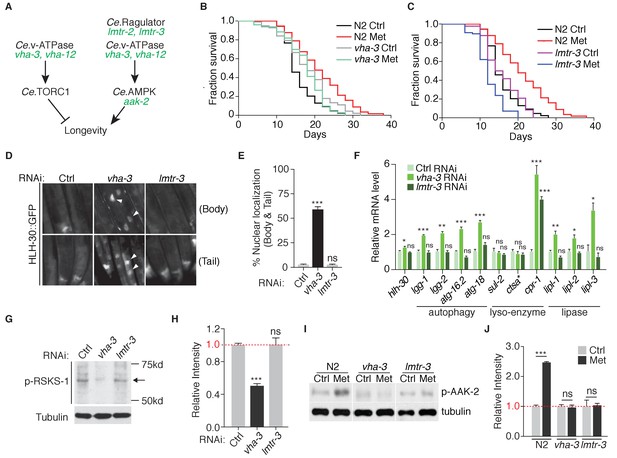
Metformin extends lifespan through v-ATPase-Ragulator-dependent activation of AMPK.
(A) A scheme depicting genes within C. elegans TORC1 and AMPK pathways. (B–C) Lifespan analysis of wild type, vha-3 (B), or lmtr-3 (C) animals in the presence or absence of metformin. (D) Representative fluorescent images of HLH-30 nuclear localization in worms administrated with control, vha-3 or lmtr-3 RNAi. (E) Quantification of (D), percentage of worms with HLH-30 nuclear accumulation. Mean ± SEM of 3 independent biological replicates are shown (sample size: n ≥ 40 worms). (F) Q-PCR analysis of HLH-30 target genes in worms administrated with control, vha-3 or lmtr-3 RNAi. ~ 300 worms were collected for each mRNA sample. Data from 3 independent biological replicates are shown as mean ± SEM. (G) Representative immunoblots of RSKS-1 phosphorylation in worms administrated with control, vha-3 or lmtr-3 RNAi. (H) Quantification of (G).~300 worms were pooled in each protein sample. Relative intensities of 3 independent biological replicates are shown as mean ± SEM. (I) Representative immunoblots of AAK-2 phosphorylation in wild type, vha-3 or lmtr-3 mutants, in the presence or absence of metformin. (J) Quantification of (I).~300 worms were collected in each protein sample. Relative intensities of 3 independent biological replicates are shown as mean ± SEM. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
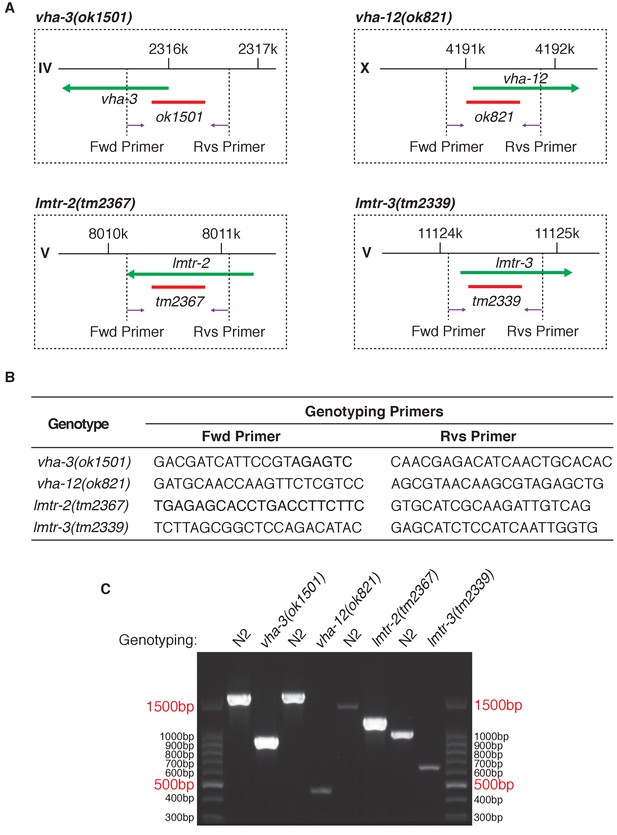
Genotyping of v-ATPase and Ragulator mutants.
(A) Schemes depicting deletion and PCR primer positions within each mutant. (B) Primer sequences for genotyping. (C) PCR results of genotyping.
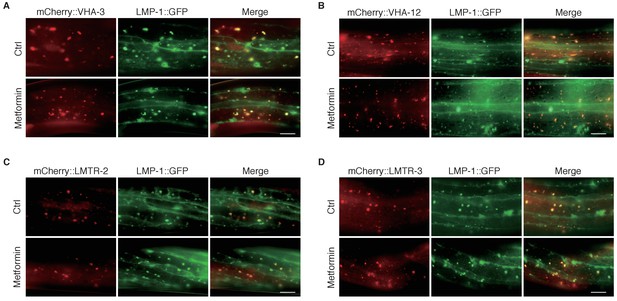
Lysosomal localization of C.elegans v-ATPase-Ragulator proteins.
(A–D) Representative fluorescent images to test co-localization of VHA-3, VHA-12, LMTR-2 or LMTR-3 with LMP-1. L1 worms were cultured in the presence or absence of metformin until L4 stage. Scale bar: 10 um.
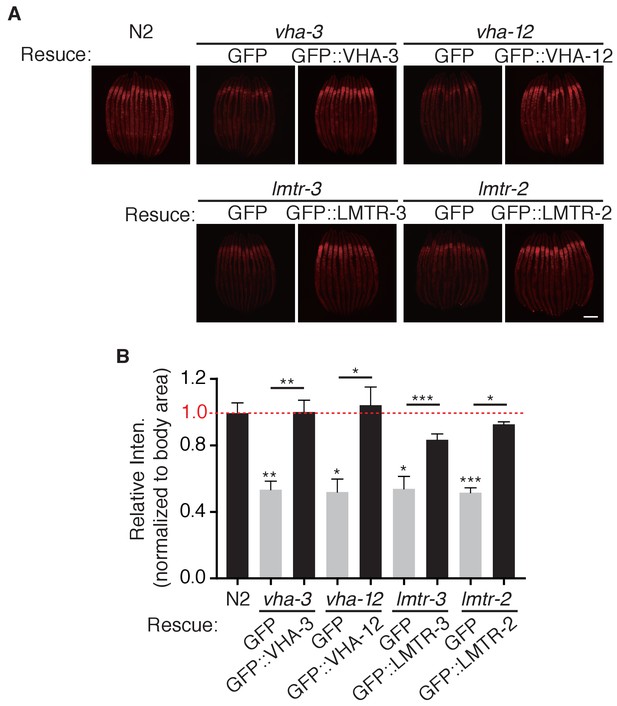
Mutation of v-ATPase or Ragulator perturbs lysosomal function in C.elegans.
(A) Representative fluorescent images of Magic Red Cathepsin assay in wild-type, vha-3, vha-12, lmtr-2, or lmtr-3 loss-of-function mutants, and vha-3, vha-12, lmtr-2, or lmtr-3 loss-of-function mutants injected with the corresponding rescue plasmid. Scale bar: 100 um. (B) Quantification of (A). Error bars represent mean ± SEM of 3 independent biological replicates (sample size: n ≥ 40). ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
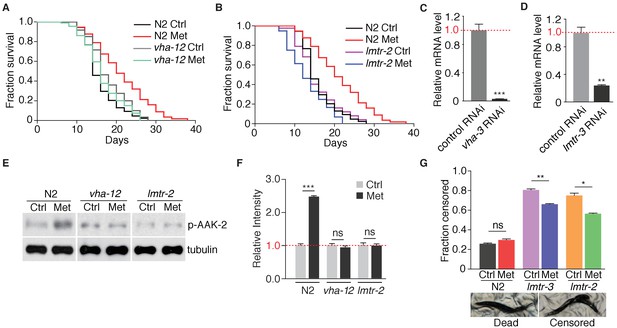
Metformin extends lifespan through v-ATPase-Ragulator-dependent AMPK activation.
(A–B) Lifespan assays of wild type, vha-12 (A), or lmtr-2 mutants (B) in the presence or absence of metformin. (C–D) Knockdown efficiency of vha-3 (C) or vha-12 RNAi (D). (E) Representative immunoblots of AAK-2 phosphorylation in N2, vha-12, or lmtr-2 mutants treated with or without metformin. (F) Quantification of (E).~300 worms were pooled in each protein sample. p-AAK-2 intensities were normalized to that of tubulin, and relative intensities of 3 independent biological replicates are shown as mean ± SEM. (G) Censored worms due to vulva blasting were counted during lifespan analysis of N2, lmtr-3 or lmtr-2 worms in the presence or absence of metformin. Error bars represent mean ± SEM of two independent biological replicates (sample size:~100 N2 worms;~250 lmtr-2/3 mutants). ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
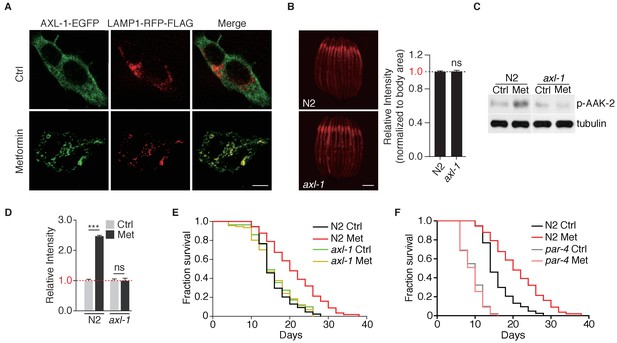
Metformin extends lifespan through axl-1-dependent activation of AMPK.
(A) Representative fluorescent images to test co-localization of AXL-1 with LAMP1. HEK293T cells stably expressing AXL-1-EGFP and LAMP1-RFP-FLAG were cultured in the presence or absence of 2 mM metformin for 12 hr. Scale bar: 10 um. (B) Representative fluorescent images of Magic Red Cathepsin assay in wild type worms or axl-1 mutants. Scale bar: 100 um. Error bars represent mean ± SEM of 3 independent biological replicates (sample size: n ≥ 40). (C) Representative immunoblots of AAK-2 phosphorylation in wild type or axl-1 mutants with or without metformin treatment. (D) Quantification of (C).~300 worms were pooled in each protein sample. Relative intensities of 3 independent biological replicates are shown as mean ± SEM. (E–F) Lifespan analysis of wild type, axl-1 (E), or par-4 mutants (F) in the presence or absence of metformin. ns, no significant difference; *p<0.05; **p<0.01; ***p<0.001.
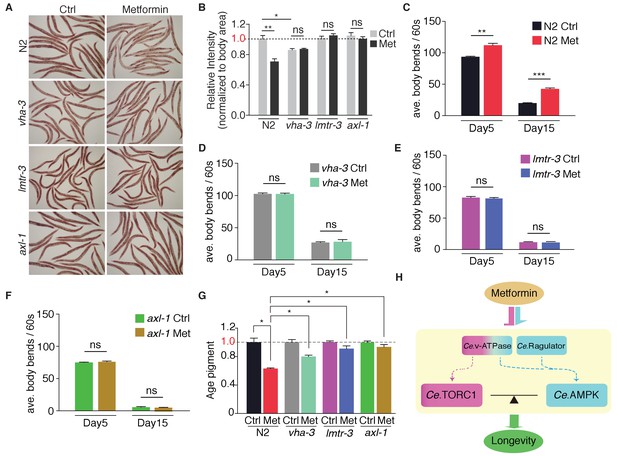
Metformin attenuates age-related fitness decline through lysosome-dependent activation of AMPK.
Neutral fat deposition (A–B), locomotion (C–F) and age pigments (G) were measured in wild-type worms, or vha-3, lmtr-3 or axl-1 mutants in the presence or absence of metformin. Error bars represent mean ± SEM of 3 independent biological replicates. (sample size: n ≥ 20 worms for locomotion assay; n ≥ 40 worms for ORO staining or age pigments assay). (H) Metformin may target v-ATPase-Ragulator complex and promote longevity through coordination of Ce.TORC1 and Ce.AMPK.
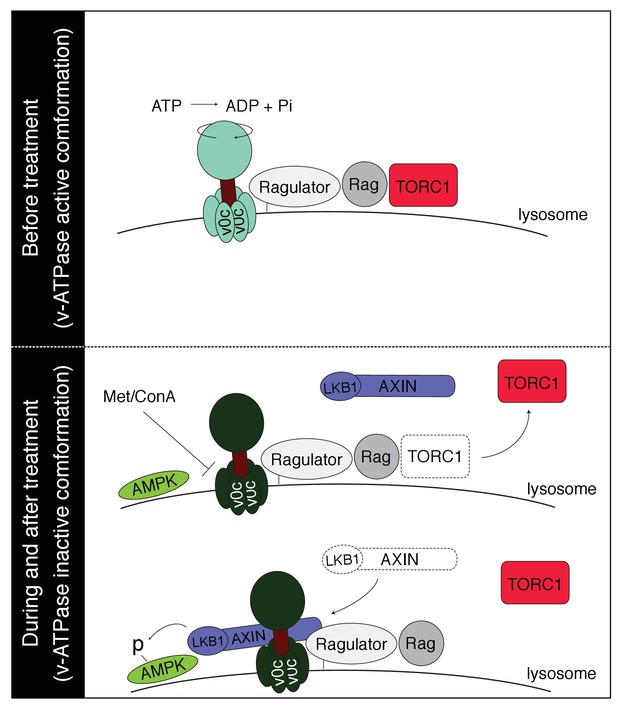
Proposed model for metformin’s action.
Under normal condition, v-ATPase is in an active conformation, which mediates lysosomal amino acid signaling to recruit and activate TORC1. Metformin treatment may target v-ATPase and change its conformation, leading to the dissociation and inactivation of TORC1. Meanwhile, v-ATPase-Regulator may act as a platform for the docking of AXIN/LKB1 and subsequent activation of AMPK.
Additional files
-
Supplementary file 1
Strains and RNAi used in this study.
- https://doi.org/10.7554/eLife.31268.021
-
Supplementary file 2
Lifespan anaylsis of metformin's effect on daf-15 heterzogous mutants.
- https://doi.org/10.7554/eLife.31268.022
-
Supplementary file 3
Lifespan analysis of metformin's effect on mutants of TORC1 downstream genes.
- https://doi.org/10.7554/eLife.31268.023
-
Supplementary file 4
Lifespan analysis of metformin's effect v-ATPase, Ragulator, AXIN/LKB1 mutants.
- https://doi.org/10.7554/eLife.31268.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31268.025





