PTEN negatively regulates the cell lineage progression from NG2+ glial progenitor to oligodendrocyte via mTOR-independent signaling
Figures
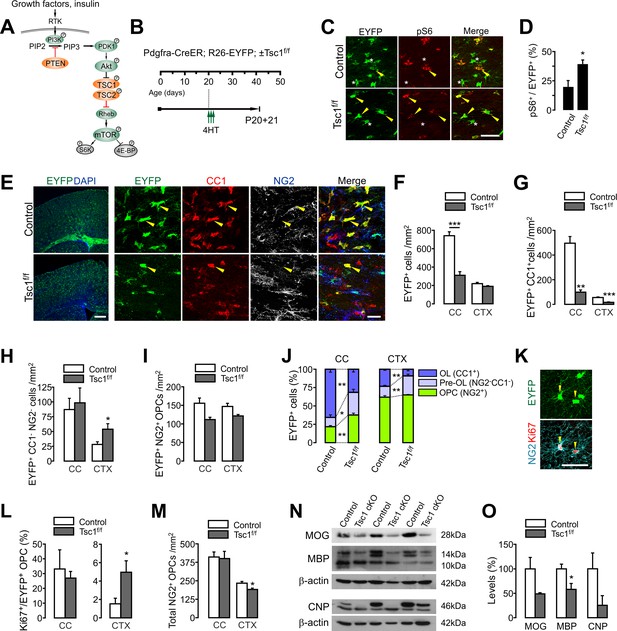
OPC-specific Tsc1 ablation impairs oligodendrocyte development in the brain.
(A) Schematic diagram of the Akt-mTOR signaling pathway. The TSC1/2 complex and PTEN (orange circles) negatively regulate mTOR activity, whereas other molecules in green circles positively regulate it. (B) Experimental scheme for 4HT administration into Pdgfra-CreER; R26-EYFP; ±Tsc1f/f mice and mouse sampling. Three 4HT injections (1 mg per injection) were given between P20 and P21 (a total of 3 mg of 4HT). (C) Confocal images of phosphorylated S6 ribosomal protein (pS6) and EYFP+ cells in the CC at P20 +21. Arrowheads and asterisks indicate EYFP+ pS6+ cells and EYFP+ pS6- cells, respectively. Scale bar, 50 µm. (D) Quantification of the percentage of pS6+ cells among EYFP+ cells in the CC. n = 4 mice per group. (E) Fluorescence (left) and confocal microscopic (right) images of EYFP+ cells in the control and Tsc1 cKO mice (P20 +21). The confocal images of EYFP+ cells were taken from the CTX, and show their maturation stages. Arrowheads indicate EYFP+CC1+ OLs. Scale bars, 500 μm (left) and 50 μm (right). (F) Quantification of EYFP+ cells in the CC and CTX. (G - I) The numbers of EYFP+CC1+ OLs (G), EYFP+CC1-NG2- pre-OLs (H), and EYFP+NG2+ OPCs (I). (J) Percentages of OPC, pre-OL and OL among EYFP-labeled cells at P20 +21. n = 6 (control) or 3 (Tsc1 cKO) mice for (F - J). (K) Cell proliferation analysis with Ki67-expressing patterns. Confocal images of EYFP+NG2+Ki67+ OPCs in the CTX of a Tsc1 cKO mouse. Scale bar, 50 μm. (L) The percentage of Ki67+ cells among EYFP+ OPCs. n = 4 mice per group. (M) The number of total OPCs. (N) Western blot analysis of cortical lysates (P20 +21) for myelin proteins MOG, MBP, and CNP. Tsc1f/f (control) and 4HT-administered Pdgfra-CreER; Tsc1f/f mice (Tsc1 cKO) were used. (O) Quantification of levels of myelin proteins. n = 3 mice per group for (N, O). Data are represented as mean ±S.E.M. *p<0.05; **p<0.01; ***p<0.001. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 1—source data 1. Original western images are available in Figure 1—source data 2.
-
Figure 1—source data 1
Numerical data for graphs in Figure 1.
- https://doi.org/10.7554/eLife.32021.005
-
Figure 1—source data 2
Original western blot images used for Figure 1K.
Full-length western blot images from two separate blots (A and B). The blots were sequentially re-probed with the indicated Abs. The original images were cut with dashed-line boxes.
- https://doi.org/10.7554/eLife.32021.006
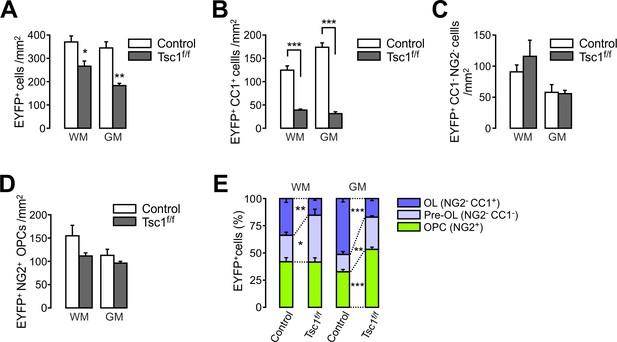
Tsc1 deletion in OPCs impairs new oligodendrocyte development in the spinal cord.
(A) The numbers of total EYFP+ cells were reduced in the ventral spinal cord (SC) of Pdgfra-CreER; R26-EYFP; Tsc1f/f (P20 +21) mice compared to their controls (Pdgfra-CreER; R26-EYFP). WM, white matter. GM, gray matter. (B - D) The number of EYFP+CC1+ OLs (B), EYFP+CC1-NG2- pre-OLs (C), and EYFP+NG2+ OPCs (D) in the SC. (E) Percentages of OPC, pre-OL, and OL among EYFP-labeled cells. Data are represented as mean ±S.E.M. n = 6 (control) or 3 (Tsc1 cKO) mice. *p<0.05; **p<0.01; ***p<0.001. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Numerical data for graphs in Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.32021.004
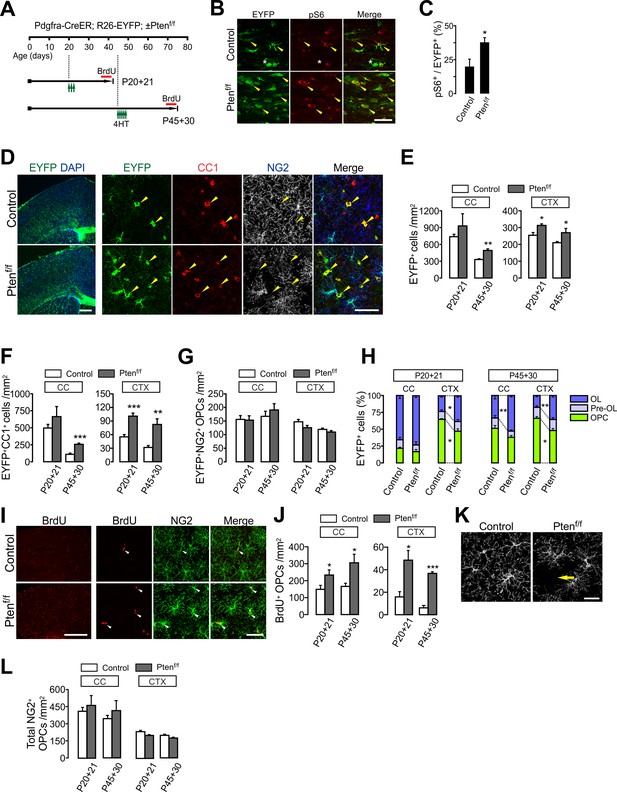
OPC-specific Pten ablation enhances oligodendrocyte differentiation and OPC proliferation in the brain.
(A) Experimental scheme for 4HT injection and BrdU administration into Pdgfra-CreER; R26-EYFP; ±Ptenf/f mice, and for mouse sampling. For P45 +30, 4HT (1 mg per injection) was injected five times between P45 and P47 (a total of 5 mg). (B) Confocal images of phosphorylated S6 ribosomal protein (pS6) and EYFP+ cells in the CC at P20 +21. Arrowheads and asterisks indicate EYFP+ pS6+ cells and EYFP+pS6- cells, respectively. Scale bar, 50 µm. (C) Percentage of pS6+ cells among EYFP-labeled cells in the CC at P20 +21. n = 5 mice per group. (D) Fluorescence (left) and confocal (right) images of EYFP+ cells in the brains of the 4HT-administered control and Pten cKO mice (P20 +21). The confocal images were taken from the CTX. Arrowheads indicate EYFP+CC1+ mature OLs. Scale bars, 500 μm (left) and 50 μm (right). (E) Number of total EYFP+ cells was increased in the CTX of Pten cKO mice during the OPC fate analysis for the two age windows. (F) Number of EYFP+CC1+ OLs. (G) The numbers of EYFP+NG2+ OPCs were not changed by the Pten cKO. (H) Percentages of OPC, pre-OL and OL among EYFP-labeled cells. (I) Fluorescence (left) and confocal (right) images of BrdU+ cells in the CTX (P20 +21). Arrowheads indicate BrdU+NG2+ OPCs. Scale bars, 500 μm (left) or 50 μm (right). (J) Quantification of BrdU+NG2+ OPCs in the CC and CTX. (K) Confocal images showing disruption of tiled OPC distribution in the CTX of Pten cKO mice (P20 +21). An arrow indicates a cortical area devoid of an NG2+ OPC. Scale bar, 50 μm. (L) Number of total OPCs. Data are represented as mean ±S.E.M. n = 4 ~ 7 mice per group for P20 ~41. n = 3 ~ 5 mice per group for P45 ~P75. *p<0.05; **p<0.01; ***p<0.001. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 2—source data 1.
-
Figure 2—source data 1
Numerical data for graphs in Figure 2.
- https://doi.org/10.7554/eLife.32021.010
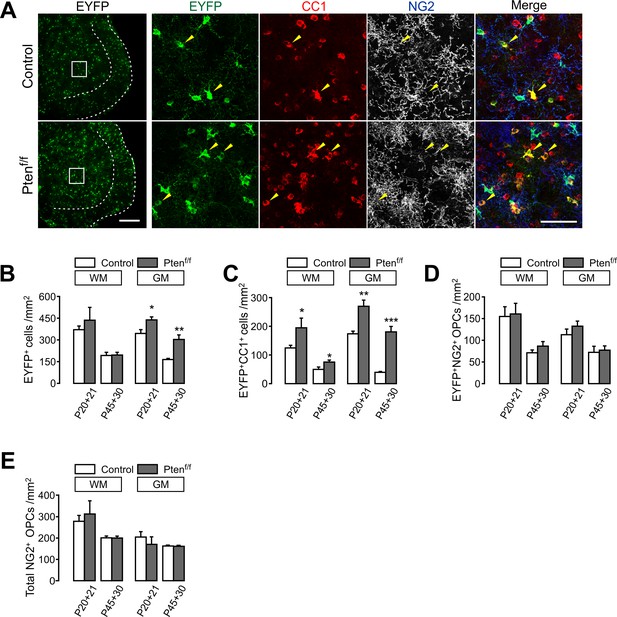
OPC-specific Pten ablation enhances oligodendrocyte differentiation in the spinal cord.
(A) Fluorescence (left) and confocal (right) images of SC EYFP+ cells in Pdgfra-CreER; R26-EYFP; ±Ptenf/f mice (P20 +21). The identity of EYFP+ cells in the boxed areas (left) was determined by co-immunostaining with maturation stage markers (right). Arrowheads indicate newly generated EYFP+CC1+ OLs. Scale bars, 100 µm (left) and 50 µm (right). (B - E) Quantification of EYFP+ cells (B), EYFP+CC1+ new OLs (C), EYFP+NG2+ OPCs (D) and total NG2+ OPCs (E) in the SC of control and Pten cKO mice. Data are represented as mean ±S.E.M. n = 3 ~ 6 (control), or 3 ~ 5 (Pten cKO) mice. *p<0.05; **p<0.01; ***p<0.001. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
Numerical data for graphs in Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.32021.009
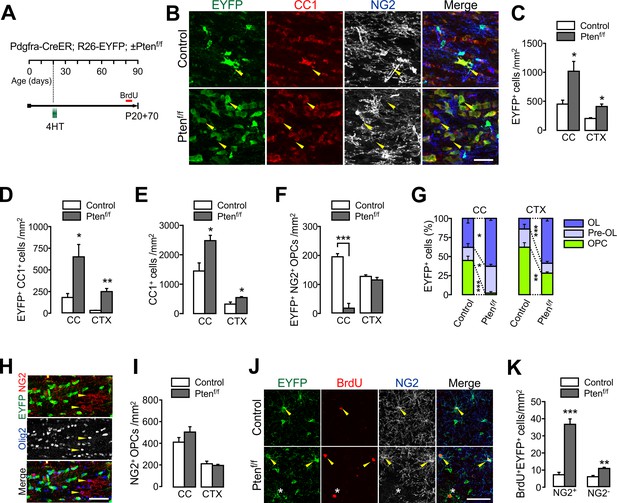
PTEN-deficient OPCs continuously add new oligodendrocytes.
(A) Experimental scheme for 4HT and BrdU administration into Pdgfra-CreER; R26-EYFP; ±Ptenf/f mice, and for mouse sampling at P90 (P20 +70). (B) Confocal images of EYFP+ cells in the CC showing their maturation stage. Arrowheads indicate EYFP+CC1+ OLs. Scale bar, 50 µm. (C) Number of EYFP+ cells in the CC and CTX at P20 +70. (D) Number of EYFP+CC1+ OLs. (E) Number of total CC1+ OLs. (F) Number of EYFP+NG2+ OPCs. (G) Percentages of OPC, pre-OL and OL among EYFP-labeled cells in the control and Pten cKO mice at P20 +70. (H) Confocal images of NG2+ OPCs in the CC of Pten cKO mice. At P20 +70, resident Olig2+NG2+ OPCs did not EYFP+ OPCs (arrowheads) in the CC. Scale bar, 50 µm. (I) The number of total NG2+ OPCs did not change in Pten cKO mice. (J) Confocal images of cortical EYFP-labeled BrdU+ cells. Arrowheads and asterisk indicate BrdU+NG2+ OPCs and a BrdU+NG2- cell, respectively. (K) Number of BrdU+EYFP+NG2+ OPCs and BrdU+EYFP+NG2- OL lineage cells in the CTX. Data are represented as mean ±S.E.M. n = 3 (control) and 4 (Pten cKO) mice. *p<0.05; **p<0.01; ***p<0.001. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 3—source data 1.
-
Figure 3—source data 1
Numerical data for graphs in Figure 3.
- https://doi.org/10.7554/eLife.32021.014
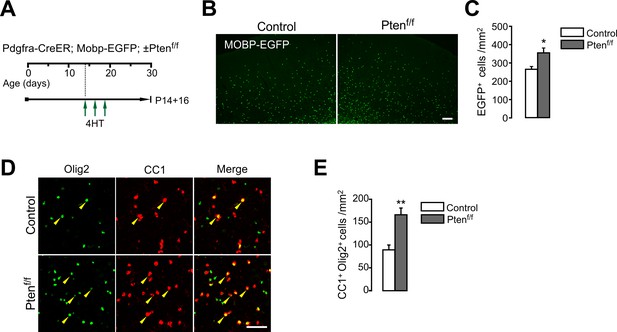
OPC-specific Pten ablation facilitates oligodendrocyte accumulation in the cortex.
(A) The experimental scheme for 4HT administration into Pdgfra-CreER; Mobp-EGFP; ±Ptenf/f mice and mouse sampling (P14 +16). 4HT was injected at P14, 16 and 18 (one single injection per day). (B) Fluorescence images of EGFP+ cells in the CTX. Scale bar, 100 µm. (C) Quantification of EGFP+ OLs in the CTX at P14 +16. (D) Confocal images of cortical Olig2+CC1+ OLs (arrowheads). Some CC1+ (or EGFP+) OLs were not labeled with our Olig2 immunostaining. Scale bar, 50 µm. (E) Quantification of Olig2+CC1+ OLs in the CTX at P14 +16. Data are represented as mean ±S.E.M. n = 4 (control) or 3 (Pten cKO) mice. *p<0.05; **p<0.01. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Numerical data for graphs in Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.32021.013
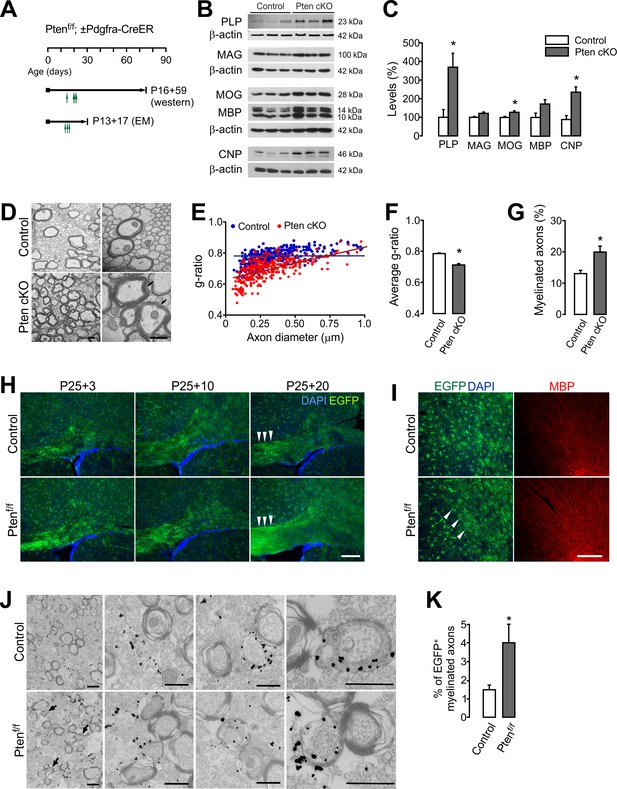
OPC-specific Pten ablation promotes new myelination.
(A) Experimental scheme for 4HT into Ptenf/f ± Pdgfra-CreER mice, and for mouse sampling for western blot analysis (B,C) and EM analyses (D –G). For western blot analysis, 4HT was injected into Ptenf/f (control) and Pdgfra-CreER; Ptenf/f mice at P14, P20 (twice), and P21 (1 mg per injection, a total of 4 mg), and the mice were sampled at P75. For EM, A single dose 4HT was injected at P13, 15, and 17 (a total of 3 mg), and the mice were sampled at P30. (B) Western blot analysis of cortical lysates for myelin proteins of control and OPC-targeted Pten cKO mice (P14 +61). (C) Quantification of the levels of PLP, MAG, MOG, MBP and CNP in the western blot (B). n = 3 mice per group. (D) Representative electron micrograph (EM) of the CC of the control and Pten cKO mice (P13 +17). Arrows indicate thickened myelin in Pten cKO mice. Scale bar, 500 nm. (E) Scatter plot of g-ratios. More than 100 myelinated axons per mouse were analyzed. (F) Average g-ratio. (G) Percentage of myelinated axons was increased in Pten cKO mice. More than 700 axons were analyzed for myelination per mouse. n = 3 mice per group for (D - G). (H, I) Fluorescence images of EGFP+ cells in the CC (G) and CTX (H) of Pdgfra-CreER; R26-mEGFP;±Ptenf/f mice. 4HT (1 mg per injection, two injections per day) was injected at P25 and P26 (a total of 4 mg), and the mice were killed 3, 10, or 20 days later. Arrowheads indicate increased EGFP+ slender processes, reminiscent of bundles of myelinated fibers in Pten cKO mice (P25 +20). Scale bar, 100 µm. (J) Immuno-EM of anti-EGFP immuno-gold particles in the CC of Pdgfra-CreER; R26-mEGFP; ±Ptenf/f mice (P20 +21). Arrows indicate EGFP+ newly formed immature myelin sheaths. Scale bar, 500 nm. (K) Percentage of EGFP+ myelinated axons increased at P20 +21. n = 3 mice per group. Data are represented as mean ±S.E.M. *p<0.05. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 4—source data 1. Original western images are available in Figure 4—source data 2.
-
Figure 4—source data 1
Numerical data for graphs in Figure 4.
- https://doi.org/10.7554/eLife.32021.016
-
Figure 4—source data 2
Original western blot images used for Figure 4A.
Full-length western blot images from three separate blots (A, B, and C) used in Figure 4A. The blots were sequentially re-probed as indicated. The original images were cut with dashed-line boxes. The blot D was used for the simultaneous detection PLP and β-actin using Odyssey infrared scanner (LI-COR).
- https://doi.org/10.7554/eLife.32021.017
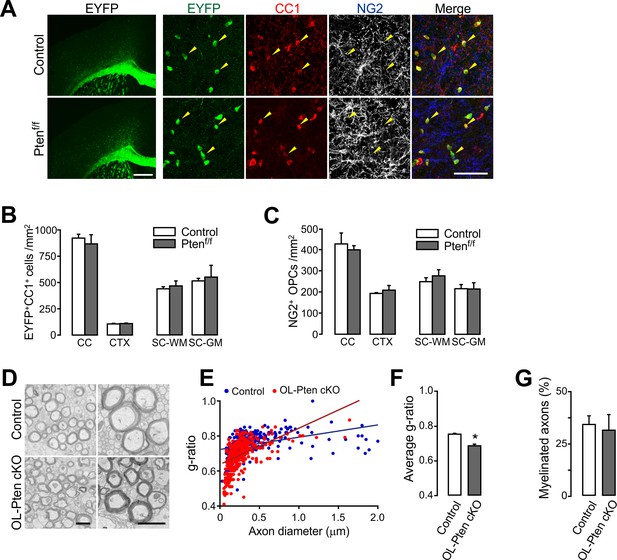
Oligodendrocyte numbers and the degree of new myelination are not changed by OL-specific Pten ablation.
(A) Fluorescence (left) and confocal (right) images of EYFP+ cells in Mog-iCre; R26-EYFP; ±Ptenf/f (P41) mice. The confocal images were taken from the CTX. Arrowheads indicate EYFP+CC1+ OLs. Scale bars, 500 µm (left) and 50 (right) µm. (B) Number of EYFP+CC1+ OLs in the brain and the SC. (C) Number of NG2+ OPCs. (D) Representative EM of myelinated callosal axons from R26-EYFP; Ptenf/f (control) and R26-EYFP; Ptenf/f; ±Mog-iCre (Pten cKO) mice (P34). Scale bar, 500 nm. (E) Scatter plot of the g-ratios. More than 100 axons per mouse, and three mice per group were analyzed. (F) Average g-ratio. (G) Percentage of myelinated axons was not altered in OL-specific Pten cKO mice. Data are represented as mean ±S.E.M. n = 3 (control) or 4 (Pten cKO) mice for (B, C), and n = 3 mice per group for (D - G). *p<0.05. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 5—source data 1.
-
Figure 5—source data 1
Numerical data for graphs in Figure 5.
- https://doi.org/10.7554/eLife.32021.019
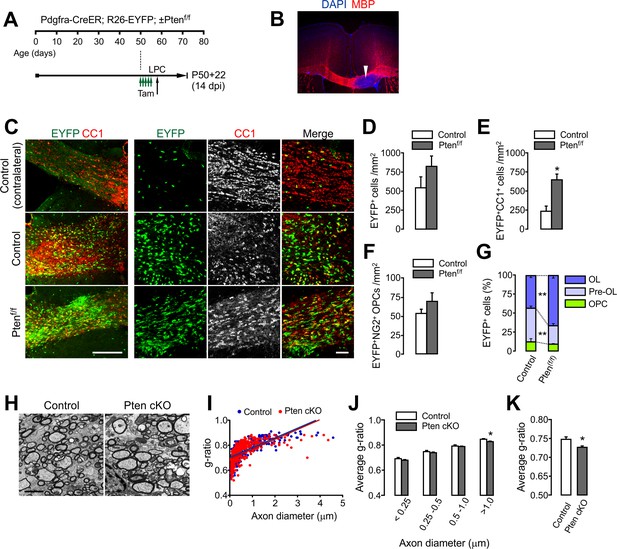
OPC-specific Pten ablation facilitates oligodendrocyte regeneration and remyelination after LPC-induced demyelination.
(A) Experimental scheme for tamoxifen (Tam) and lysolecithin (LPC) injection into Pdgfra-CreER; R26-EYFP; ±Ptenf/f mice. A series of tamoxifen injections (40 mg/kg per i.p. injection, a total of 10 injections) were given between P50 and P54. The demyelination was induced with LPC injection at P58, and the mice were sampled at P72, which is 14 days after LPC injection (14 dpi), and 22 days after the first tamoxifen injection (P50 +22). (B) Loss of MBP immunoreactivity (arrowhead) at the LPC-injected CC in the brain. (C) Fluorescence (left) and confocal (right) images of EYFP+ cells and CC1+ OLs in the CC at 14 days after LPC injection (14 dpi). Scale bars, 200 μm (left) and 50 µm (right). (D) Quantification of EYFP+ cells in the LPC injected site at 14 dpi. (E) Quantification of EYFP+CC1+ OLs. (F) Quantification of EYFP+NG2+ OPCs. (G) Pten cKO mice exhibited marked changes in the percentage of OL and pre-OL among EYFP-labeled cells at the LPC-injected areas at 14 dpi. (H) Representative EMs of the lesion in the CC at 14 dpi. Scale bar, 2 μm. (I) Scatter plot for individual g-ratios of myelinated axons. More than 100 myelinated axons per mouse, and three mice per group were analyzed. (J, K) Average g-ratio according to axon diameter (J) and as a total (K). Data are represented as mean ±S.E.M. n = 3 (control) or 5 (Pten cKO) mice for (D - G), or n = 3 per group for (H–K). *p<0.05; **p<0.01. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 6—source data 1.
-
Figure 6—source data 1
Numerical data for graphs in Figure 6.
- https://doi.org/10.7554/eLife.32021.023
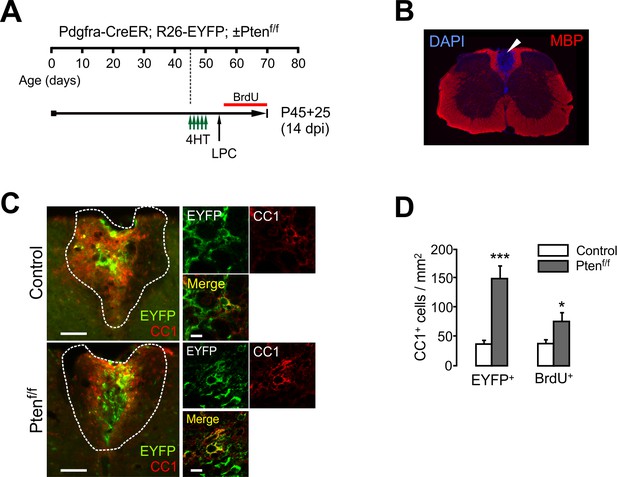
OPC-specific Pten ablation facilitates oligodendrocyte regeneration after lysolecithin-induced demyelination in the spinal cord.
(A) Experimental scheme for 4HT, BrdU and LPC injection into Pdgfra-CreER; R26-EYFP; ±Ptenf/f mice, and for mouse sampling. Five injections of 4HT (1 mg per injection) were given between P45 and P47, and LPC was injected at P55. BrdU was administered from P56 to the sampling age only via drinking water. (B) Loss of MBP immunoreactivity (arrowhead) at an LPC-injected area in the SC dorsal WM. (C) Fluorescence (left) and confocal images (right) of the SC dorsal WM at 14 dpi. Scale bars, 100 µm (left) and 10 µm (right). (D) Quantification of newly generated EYFP+CC1+ OLs (left) and BrdU+CC1+ cells (right). Data are represented as mean ±S.E.M. n = 3 mice per group. *p<0.05; ***p<0.001. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 6—figure supplement 1—source data 1.
-
Figure 6—figure supplement 1—source data 1
Numerical data for graphs in Figure 6—figure supplement 1.
- https://doi.org/10.7554/eLife.32021.022
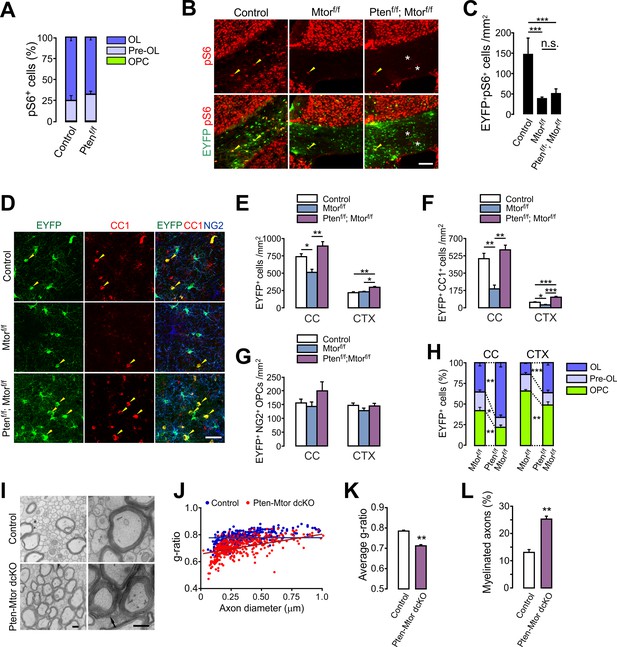
mTOR is dispensable for the enhanced oligodendrocyte generation from PTEN-deficient OPCs and the subsequent hypermyelination.
(A) Percentage of OPC, pre-OL and OL among EYFP+pS6+ cells in the CC of the control (Pdgfra-CreER; R26-EYFP) and Pten cKO (Pdgfra-CreER; R26-EYFP; Ptenf/f) mice (P20 +21). Almost all pS6 immunoreactivities were observed either in EYFP+NG2- pre-OLs or in EYFP+CC1+ OLs, but not in NG2+ OPCs. More than 150 callosal EYFP+pS6+ cells were analyzed per mouse. n = 4 mice per group. (B) Validation of effective mTOR inactivation with pS6 immunofluorescence in OPC-specific Mtor and Pten-Mtor double cKO mice (Pdgfra-CreER; R26-EYFP; ±Ptenf/f; ±Mtorf/f, P20 +21). Arrowheads and asterisks indicate pS6+EYFP+ and pS6+EYFP- cells, respectively. Scale bar, 100 µm. (C) Quantification of the EYFP+pS6+ cells in the CC. n = 3 mice per group. (D) Confocal images of EYFP+ cells in the CTX showing their maturation stage. Arrowheads indicate newly differentiated EYFP+CC1+ OLs. Scale bar, 50 µm. (E - G) Quantification of total EYFP+ cells (E), EYFP+CC1+ OLs (F), and EYFP+NG2+ OPCs (G) in the CC. (H) Percentages of OPC, pre-OL and OL among EYFP-labeled cells of Mtor cKO and Pten-Mtor cKO mice (P20 +21). (I) Representative EM of callosal axons in the control (Ptenf/f) and Pten-Mtor cKO mice (P13 +17). An arrow indicates altered myelin thickness. Scale bar, 500 nm. (J) Scatter plot of individual g-ratios. (K) Average of g-ratios. At least 130 axons per mouse, and three mice per group were analyzed. (L) Percentage of myelinated axons. Data are represented as mean ±S.E.M. n = 6 (control), 4 (Mtor cKO), or 4 (Pten-Mtor cKO) mice for (E - H), n = 3 mice per group for (I - L). *p<0.05; **p<0.01; ***p<0.001. One-way ANOVA with Bonferroni test for (C, E – G). Unpaired Student's t-test for (H, K, L). The numerical data for the graphs are available in Figure 7—source data 1.
-
Figure 7—source data 1
Numerical data for graphs in Figure 7.
- https://doi.org/10.7554/eLife.32021.029
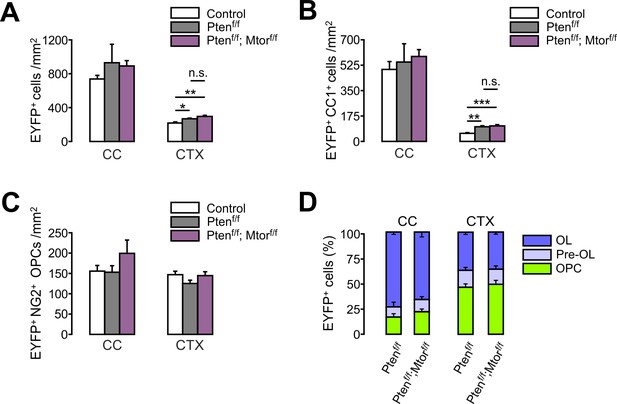
Additional Mtor ablation does not reverse Pten-ablation-induced oligodendrocyte promotion in the brain.
(A) Quantification of EYFP+ cells in the control (Pdgfra-CreER; R26-EYFP), Pten cKO, and Pten-Mtor double cKO mice (P20 +21). (B, C) Quantification of EYFP+CC1+ OLs (B) and EYFP+NG2+ OPCs (C). (D) Percentages of OPC, pre-OL, and OL among EYFP-labeled cells. Data are represented as mean ±S.E.M. n = 4 (control and Pten-Mtor double cKO) or 5 (Pten cKO) mice. One-way ANOVA with Bonferroni post hoc test for (A - C), and unpaired Student's t-test for (D). The numerical data for the graphs are available in Figure 7—figure supplement 1—source data 1.
-
Figure 7—figure supplement 1—source data 1
Numerical data for graphs in Figure 7—figure supplement 1.
- https://doi.org/10.7554/eLife.32021.026
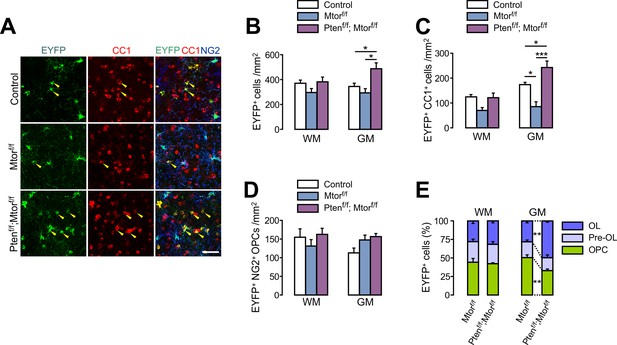
Additional Pten ablation restores oligodendrocyte differentiation in the spinal cord of Mtor cKO mice.
(A) Confocal images of EYFP+ cells in the GM-SC of the control (Pdgfra-CreER; R26-EYFP), Mtor cKO, and Pten-Mtor double cKO mice. Arrowheads indicate newly generated EYFP+CC1+ OLs. Scale bar, 50 µm. (B - D) Quantification of EYFP+ cells (B), EYFP+ CC1+ OLs (C), and EYFP+NG2+ OPCs (D) in the SC of control, Mtor cKO, and Pten-Mtor double cKO mice. (E) Percentages of OPC, pre-OL, and OL among EYFP-labeled cells in the SC of Mtor cKO and Pten-Mtor double cKO mice. Data are represented as mean ±S.E.M. n = 6 (control) or 4 (Mtor cKO and Pten-Mtor cKO) mice. One-way ANOVA with Bonferroni test for (B - D). Unpaired Student's t-test for (E). *p<0.05, **p<0.01, ***p<0.001. The numerical data for the graphs are available in Figure 7—figure supplement 2—source data 1.
-
Figure 7—figure supplement 2—source data 1
Numerical data for graphs in Figure 7—figure supplement 2.
- https://doi.org/10.7554/eLife.32021.028
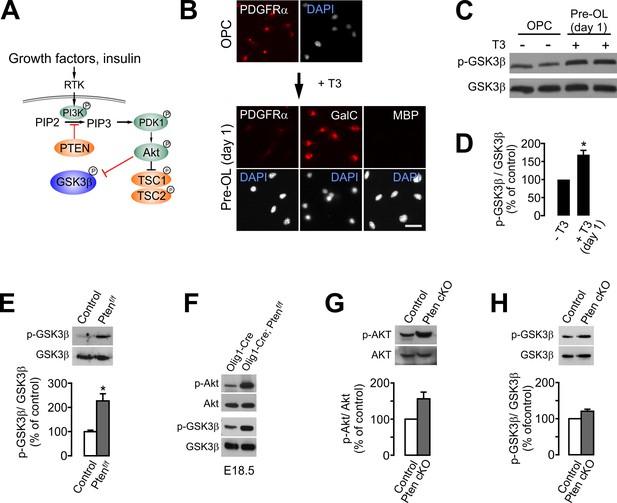
OPC-specific Pten ablation enhances the inhibitory phosphorylation (Ser9) of GSK3β.
(A) Schematic diagram for the signaling flow of PTEN-Akt-GSK3β. (B) In vitro OL differentiation from OPCs with T3. One day after T3 addition, the majority of cells become pre-OLs expressing GalC, but not MBP. Scale bar, 20 μm. (C) Western blot analysis for phospho-GSK3β (p-GSK3βSer9) levels with oligodendroglial primary culture (with or without T3 incubation for 1 day). (D) Quantification of p-GSK3βSer9 levels. n = 3 independent replicates. (E) Western blot and quantification of p-GSK3β levels in the EYFP+ cells obtained from Pdgfra-CreER; R26-EYFP; ±Ptenf/f mice (P20 +21) by FACS. n = 3 mice per group. (F) Western blot of the cortical lysates of Olig1-Cre (control) and Olig1-Cre; Ptenf/f embryo (E18.5). n = 1 mouse per group. (G, H) Western blot analyses and quantification of the levels of p-AktSer473 (G) and p-GSK3βSer9 (H) in Pten-deleted OPCs in vitro. OPC primary culture was obtained from 4HT-administered Ptenf/f (control) or Pdgfra-CreER; Ptenf/f (Pten cKO) pups (P1 +1). n = 3 for (G, H) independent replicates. Data are represented as mean ±S.E.M. *p<0.05. Paired Student's t-test for (D, G, H). Unpaired Student's t-test for (E). The numerical data for the graphs are available in Figure 8—source data 1. Original western images are available in Figure 8—source data 2.
-
Figure 8—source data 1
Numerical data for graphs in Figure 8.
- https://doi.org/10.7554/eLife.32021.031
-
Figure 8—source data 2
Original western blot images used for Figure 8E–H.
Full-length western blot images from three separate blots (A, B, and C) used in Figure 8E–H. The blots were stripped and sequentially re-probed as indicated. The original images were cut with dashed-line boxes.
- https://doi.org/10.7554/eLife.32021.032
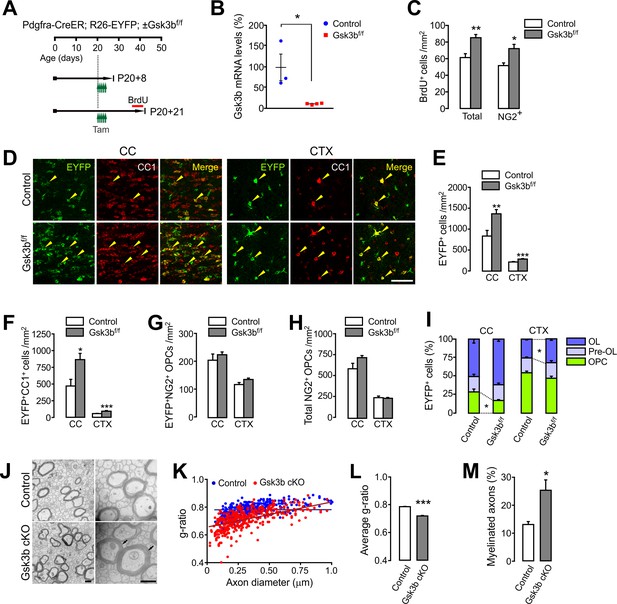
OPC-specific Gsk3b ablation enhances oligodendrocyte differentiation in the brain.
(A) Experimental scheme for tamoxifen and BrdU administration into Pdgfra-CreER; R26-EYFP; ±Gsk3bf/f and for mouse sampling. Tamoxifen (40 mg/kg per i.p. injection) was injected five times for 2.5 days starting P20. (B) Validation of Gsk3b deletion. EYFP+ cells were isolated from the CTX of Pdgfra-CreER; R26-EYFP; ±Gsk3bf/f mice by FACS at P20 +8. The isolated EYFP-labeled cells were subjected to RT-qPCR for Gsk3b mRNA levels. n = 3 (control) and 4 (Gsk3b cKO) mice. (C) Quantification of BrdU+ cells in the CTX at P20 +21. There was an increases in BrdU+NG2+ OPCs in the Gsk3b cKO mice (P20 +21). n = 5 mice per group. (D) Confocal images of EYFP+CC1+ OLs (arrowheads) in the CC and CTX. Scale bar, 50 μm. (E - H) Quantification of total EYFP+ cells (E), EYFP+CC1+ OLs (F), EYFP+NG2+ OPCs (G), and total NG2+ OPCs (H). (I) Percentages of OPC, pre-OL and OL among EYFP-labeled cells. n = 8 (control) or 6 (Gsk3b cKO) mice for (E – I). (J) Representative EM of callosal axons (P13 +17). R26-EYFP (control) or Pdgfra-CreER; R26-EYFP; Gsk3bf/f (Gsk3b cKO) mice were used. Scale bar, 500 nm. (K) Scatter plot of the g-ratios. (L) Average of g-ratio. At least 130 axons per mouse, three mice per group were analyzed. (M) Percentage of myelinated axons. Data are represented as mean ±S.E.M. n = 3 mice per group (K – M). *p<0.05; **p<0.01; ***p<0.001. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 9—source data 1.
-
Figure 9—source data 1
Numerical data for graphs in Figure 9.
- https://doi.org/10.7554/eLife.32021.037
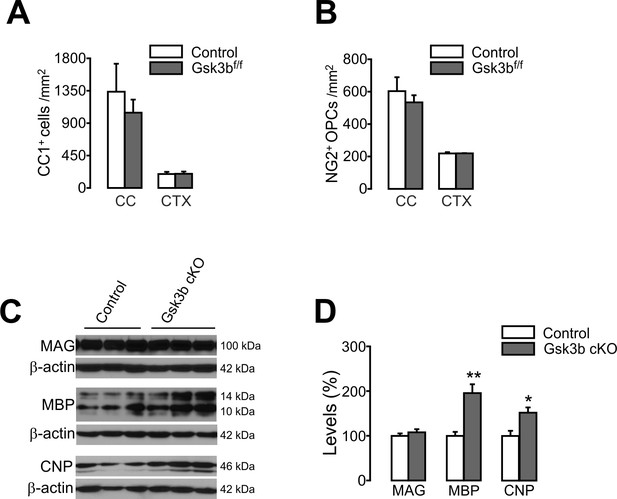
OL-specific Gsk3b ablation does not change the number of oligodendrocyte, but enhances expression levels of MBP and CNP.
(A) Quantification of CC1+ OLs in the CTX of the control (Mog-iCre; R26-EYFP) and OL-specific Gsk3b cKO (Mog-iCre; R26-EYFP; Gsk3bf/f) mice at P31. (B) Quantification of NG2+ OPCs. n = 3 mice per group for (A, B). (C, D) Western blot analysis of cortical lysates for myelin proteins, MAG, MBP, and CNP. Cortices were isolated from Gsk3bf/f (control) and Mog-iCre; Gsk3bf/f (Gsk3b cKO) mice (P31). n = 4 mice per group for (C, D). Data are represented as mean ±S.E.M. *p<0.05, **p<0.01. Unpaired Student's t-test. The numerical data for the graphs are available in Figure 9—figure supplement 1—source data 1. Original western images are available in Figure 9—figure supplement 1—source data 2. List of source data files.
-
Figure 9—figure supplement 1—source data 1
Numerical data for graphs in Figure 9—figure supplement 1.
- https://doi.org/10.7554/eLife.32021.035
-
Figure 9—figure supplement 1—source data 2
Original western blot images used for Figure 9—figure supplement 1C,D.
Full-length western blot images from three separate blots (A, B, and C). The blots were re-probed with β-actin Ab as indicated. The original images were cut as indicated with dashed-line boxes. The asterisk indicates non-specific signals.
- https://doi.org/10.7554/eLife.32021.036
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Pdgfra-CreER; Tg (Pdgfra-cre/ERT) | JAX #018280; PMID:21092857 | RRID:IMSR_JAX:018280 | |
| Genetic reagent (M. musculus) | Mog-iCre | PMID:15908920 | NA | |
| Genetic reagent (M. musculus) | Olig1-Cre; Olig1wt/Cre | JAX #011105; PMID:11955448 | RRID:IMSR_JAX:011105 | |
| Genetic reagent (M. musculus) | R26-EYFP | JAX #006148; PMID:11299042 | RRID:IMSR_JAX:006148 | |
| Genetic reagent (M. musculus) | R26-mEGFP | JAX #007576; PMID:17868096 | RRID:IMSR_JAX:007576 | |
| Genetic reagent (M. musculus) | Mobp-EGFP; Tg (Mobp-EGFP) | MMRRC (030483-UCD); PMID:23542689 | RRID:MMRRC_030483-UCD | |
| Genetic reagent (M. musculus) | Tsc1f/f | JAX #005680; PMID:12205640 | RRID:IMSR_JAX:005680 | |
| Genetic reagent (M. musculus) | Ptenf/f | JAX #006440; PMID:11857804 | RRID:IMSR_JAX:006440 | |
| Genetic reagent (M. musculus) | Mtorf/f | JAX #011009; PMID:20008564 | RRID:IMSR_JAX:011009 | |
| Genetic reagent (M. musculus) | Gsk3bf/f | JAX #029592; PMID:18694957 | RRID:IMSR_JAX:029592 | |
| Antibody | anti-Akt (rabbit monoclonal) | Cell Signaling Technology, Danvers, MA | Cat #4691; RRID:AB_915783 | dilution (1:2,000) |
| Antibody | anti-Phospho-Akt (Ser473) (rabbit monoclonal) | Cell Signaling Technology | Cat #4060; RRID:AB_2315049 | dilution (1:2,000) |
| Antibody | anti-β-actin (mouse monoclonal) | Santa Cruz Biotechnology, Dallas, TX | Cat #sc-47778; RRID:AB_2714189 | dilution (1:2,000) |
| Antibody | anti-BrdU (rat monoclonal) | Accurate, Westbury, NY | Cat #OBT0030G; RRID:AB_609567 | dilution (1:500) |
| Antibody | anti-CC1 (APC) (mouse monoclonal) | EMD Millipore, Burlington, MA | Cat #OP80; RRID:AB_2057371 | dilution (1:70) |
| Antibody | anti-CNPase (rabbit polyclonal) | PhosphoSolutions, Aurora, CO | Cat #325-CNP; RRID:AB_2492062 | dilution (1:1,000) |
| Antibody | anti-GalC (mouse monoclonal) | EMD Millipore | Cat #MAB342; RRID:AB_2073708 | dilution (1:100) |
| Antibody | anti-GFP (goat polyclonal) | Frontier Institute, Japan | Cat #GFP-Go-Af1480; RRID:AB_2571574 | dilution (1:500) |
| Antibody | anti GFP (rabbit polyclonal) | Frontier Institute | Cat #GFP-Rb-Af2020; RRID:AB_2491093 | dilution (1:500) |
| Antibody | anti-GFP (rabbit polyclonal) | Proteintech, Chicago, IL | Cat #50430–2-AP; RRID:AB_11042881 | dilution (1:500) |
| Antibody | anti-GSK3b (rabbit monoclonal) | Cell Signaling Technology | Cat #12456 | dilution (1:2,000) |
| Antibody | anti-Phospho-GSK3β (Ser9) (rabbit monoclonal) | Cell Signaling Technology | Cat #9323; RRID:AB_2115201 | dilution (1:2,000) |
| Antibody | anti-Ki67 (rabbit polyclonal) | Abcam, Cambridge, MA | Cat #ab15580; RRID:AB_443209 | dilution (1:500) |
| Antibody | anti-MAG (mouse monoclonal) | Santa Cruz Biotechnology | Cat #sc-166849; RRID:AB_2250078 | dilution (1:2,000) |
| Antibody | anti-MBP (rabbit monoclonal) | Cell Signaling Technology | Cat #78896 | dilution (1:1,000) |
| Antibody | anti-MBP (mouse monoclonal) | Covance, Princeton, NJ | Cat #SMI-99P; RRID:AB_10120130 | dilution (1:500) |
| Antibody | anti-MOG (mouse monoclonal) | Santa Cruz Biotechnology | Cat #sc-166172; RRID:AB_2145540 | dilution (1:1,000) |
| Antibody | anti-NG2 (guinea pig polyclonal) | Gift from Dr. Dwight Bergles (Johns Hopkins) | N/A | dilution (1:4,000) |
| Antibody | anti-Olig2 (rabbit polyclonal) | EMD Millipore | Cat #AB9610; RRID:AB_570666 | dilution (1:500) |
| Antibody | anti-CD140a (PDGFRα) (Clone APA5) (rat monoclonal) | BD Biosciences, San Jose, CA | Cat #558774; RRID:AB_397117 | dilution (1:250) |
| Antibody | anti-PLP (mouse monoclonal) | EMD Millipore | Cat #MAB388-100UG; RRID:AB_177623 | dilution (1:1,000) |
| Antibody | Phospho-S6 ribosomal protein (Ser240/244) (rabbit polyclonal) | Cell Signaling Technology | Cat #2215; RRID:AB_331683 | dilution (1:500) |
| Antibody | Alexa488-conjugated anti-rabbit | Jackson ImmunoResearch, West Grove, PA | Cat #703-545-152 | dilution (1:500) |
| Antibody | Alexa488-conjugated anti-goat | Jackson ImmunoResearch | Cat #705-545-147 | dilution (1:500) |
| Antibody | Cy3-conjugated anti-mouse | Jackson ImmunoResearch | Cat #715-165-151 | dilution (1:500) |
| Antibody | Cy3-conjugated anti-rabbit | Jackson ImmunoResearch | Cat #711-165-152 | dilution (1:500) |
| Antibody | Cy3-conjugated anti-rat | Jackson ImmunoResearch | Cat #712-165-153 | dilution (1:500) |
| Antibody | Alexa647-conjugated anti-guinea pig | Jackson ImmunoResearch | Cat #706-605-148 | dilution (1:500) |
| Antibody | Cy5-conjugated anti-rat | Jackson ImmunoResearch | Cat #712-175-153 | dilution (1:500) |
| Antibody | HRP-conjugated anti-mouse | Jackson ImmunoResearch | Cat #715-035-150 | dilution (1:20,000) |
| Antibody | HRP-conjugated anti-rabbit | Jackson ImmunoResearch | Cat #711-035-152 | dilution (1:20,000) |
| Antibody | IR-Dye 800 CW anti-mouse | LI-COR, Lincoln, NE | Cat #926–32210 | dilution (1:5,000) |
| Antibody | Nanogold-IgG anti-rabbit | Nanoprobes, Yaphank, NY | Cat #2003 | dilution (1:100) |
| Sequence-based reagent | Gsk3b Ex1 (F) | this paper | 5’ GACCGAGAACCACCTCCTTT 3’ | |
| Sequence-based reagent | Gsk3b Ex2 (R) | this paper | 5’ ACTGACTTCCTGTGGCCTGT 3’ | |
| Sequence-based reagent | β-actin (F) | this paper | 5’ TGACAGGATGCAGAAGGAGA 3’ | |
| Sequence-based reagent | β-actin (R) | this paper | 5' CGCTCAGGAGGAGCAATG 3’ | |
| Commercial assay or kit | TSA (+) Cyanine 3 and Fluorescein System | PerkinElmer, Hopkinton, MA | Cat #NEL753001KT | |
| Commercial assay or kit | SuperSignal West Dura | Thermo-Fisher Scientific, Waltham, MA | Cat #34075 | |
| Commercial assay or kit | Neuronal Tissue Dissociation Kit (P) | Myltenyi Biotec, Auburn, CA | Cat #130-092-628 | |
| Commercial assay or kit | RNeasy Plus Micro Kit (50) | Qiagen, Germantown, MD | Cat #74034 | |
| Commercial assay or kit | SuperScript III First-Strand synthesis system | Thermo-Fisher Scientific | Cat #18080–051 | |
| Commercial assay or kit | Pierce BCA protein assay | Thermo-Fisher Scientific | Cat #23227 | |
| Chemical compound, drug | 4-Hydroxytamoxifen | Sigma, St. Louis, MO | Cat #H-7904 | |
| Chemical compound, drug | Tamoxifen | Sigma | Cat #T-5648 | 40 mg/kg bw per injection |
| Chemical compound, drug | Odyssey Blocking Buffer (PBS) | LI-COR | Cat #927–40000 | |
| Chemical compound, drug | 5-Bromo-2'-deoxyuridine | Thermo-Fisher Scientific | Cat #BP-2508–5 | |
| Chemical compound, drug | QuantiTect SYBR | Qiagen | Cat #204143 | |
| Software, algorithm | GraphPad Prism 5.0 | GraphPad Software, La Jolla, CA | ||
| Software, algorithm | Image J | NIH, Bethesda, MD | ||
| Software, algorithm | StepOne software 2.1 | Thermo-Fisher Scientific | ||
| Software, algorithm | Image studio 3.1 | LI-COR |





