LPCAT1 controls phosphate homeostasis in a zinc-dependent manner
Figures
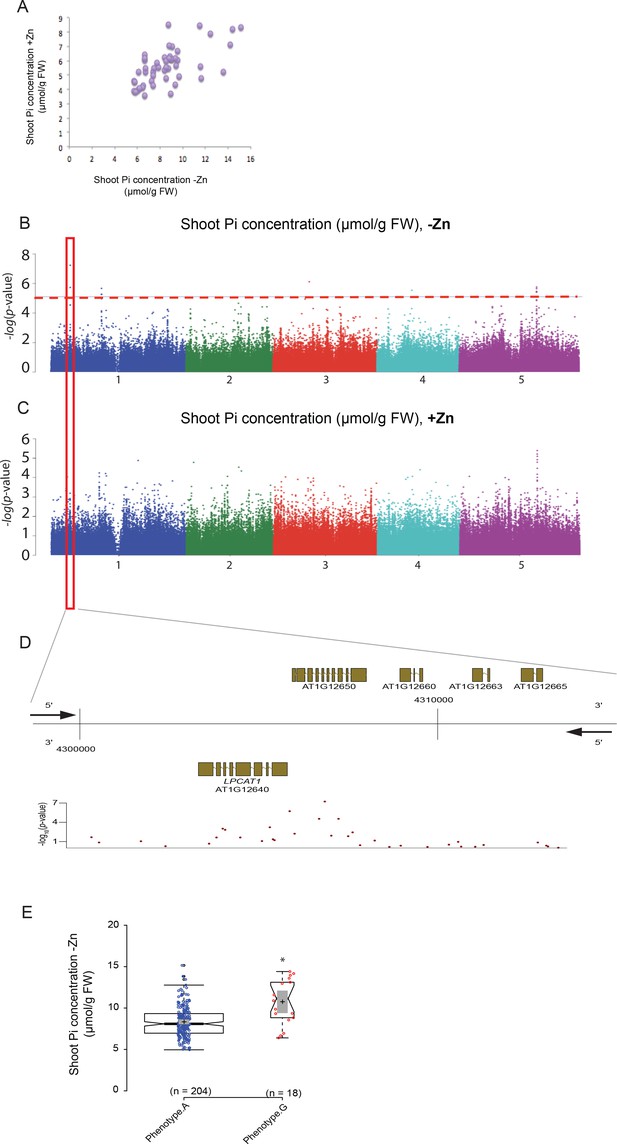
Genome-wide association (GWA) analysis of Arabidopsis shoot Pi concentration.
223 Arabidopsis thaliana accessions were grown supplemented with zinc (+Zn) or without zinc (-Zn) for 18 days under long day conditions, upon which shoot inorganic phosphate (Pi) concentrations were determined. (A) Mean shoot Pi concentration of Arabidopsis accessions in +Zn is plotted against the mean shoot Pi concentration of Arabidopsis accessions in -Zn. (B, C) Manhattan plots of GWA analysis of Arabidopsis shoot Pi concentration in -Zn (B) and +Zn (C). The five Arabidopsis chromosomes are indicated in different colours. Each dot represents the –log10(P) association score of one single nucleotide polymorphism (SNP). The dashed red line denotes an approximate false discovery rate 10% threshold. Boxes indicate the location of the LPCAT1 (red) quantitative trait loci (QTL). (D) Gene models (upper panel) and SNP –log10(P) scores (lower panel) in the genomic region surrounding the GWA QTL at the LPCAT1 (E); 5’ and 3’ indicate the different genomic DNA strands and orientation of the respective gene models. (E) Distribution of Pi concentrations in accessions with the phenotype A versus accessions with phenotype G. Asterisk indicates a significant difference between the two groups of accessions of p<0.01.
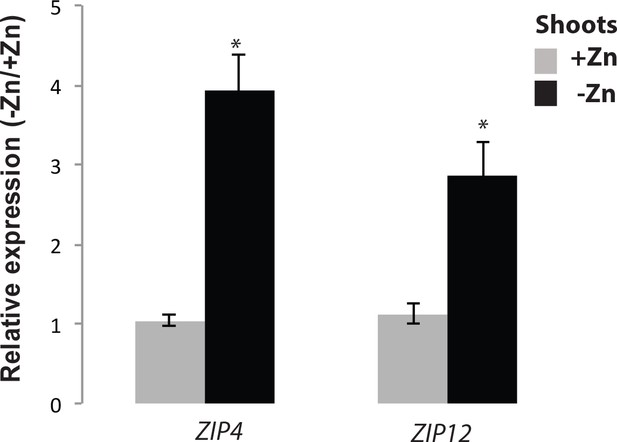
mRNA abundance of Zn-responsive genes ZIP4 and ZIP12 in roots of Col-0 plants grown in presence and absence of Zn.
Reverse transcriptase qPCR analyses of transcript level changes in response to Zn-deficiency of the genes ZIP4 (At1g10970) and ZIP12 (At5g62160) in shoots of Arabidopsis (Col-0). Seedlings were grown on vertical agar plate in presence or absence of Zn for 18 days. Transcript levels of these genes are expressed relative to the average transcript abundance of the UBIQUITIN10 (UBQ10; At4g05320) that was used as an internal control. Every data point was obtained from the analysis of shoots collected from a pool of six plants. Data presented are means of three biological replicates ±SE. Asterisks indicate statistically significant differences compared to the +Zn condition for each gene analyzed (p<0.05).
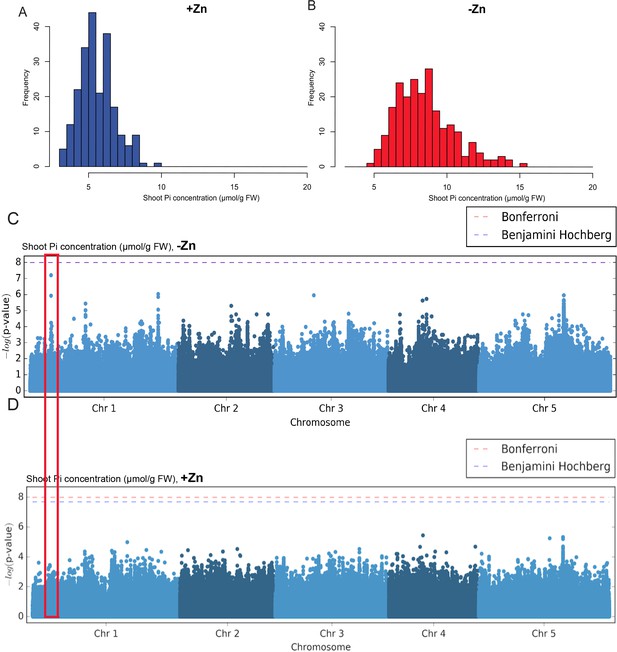
Genome-wide association (GWA) analysis of Arabidopsis shoot Pi concentration.
223 Arabidopsis thaliana accessions were grown supplemented with zinc (+Zn) or without zinc (-Zn) for 18 days under long day conditions, upon which shoot inorganic phosphate (Pi) concentrations were determined. (A, B) Histogram of the frequency distribution of mean shoot Pi concentration of Arabidopsis accessions in +Zn (A) and -Zn (B). (C, D) Manhattan plots of GWA analysis of Arabidopsis shoot Pi concentration in -Zn (C) and +Zn (D) generated using 1001 Genomes imputed SNPs. Each dot represents the –log10(P) association score of one single nucleotide polymorphism (SNP). Boxes indicate the location of the LPCAT1 (red) quantitative trait loci (QTL).
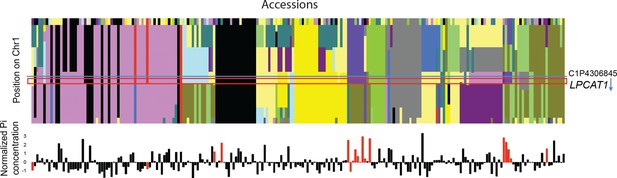
Haplotype analysis of region around SNP C1P4306845.
(upper panel) Clusterplot represents the haplotype blocks according to fastphase2 for SNPs 25 kb up and downstream of the significantly associated SNP. Colours represent a specific haplotype. X-axis: Accessions, Y-axis: SNPs. The horizontal blue line indicates the position of the significant SNP. The red box indicates the LPCAT gene model. The blue arrow represents the transcription direction of the gene model (LPCAT is on the - strand). (lower panel) Normalized Pi content in -Zn condition of each accession (using the R scale function). Colour indicates the genotype of the accession at SNP C1P4306845 (top GWAS hit), whereby red colour indicates the minor SNP allele that is associated with high Pi content in -Zn.
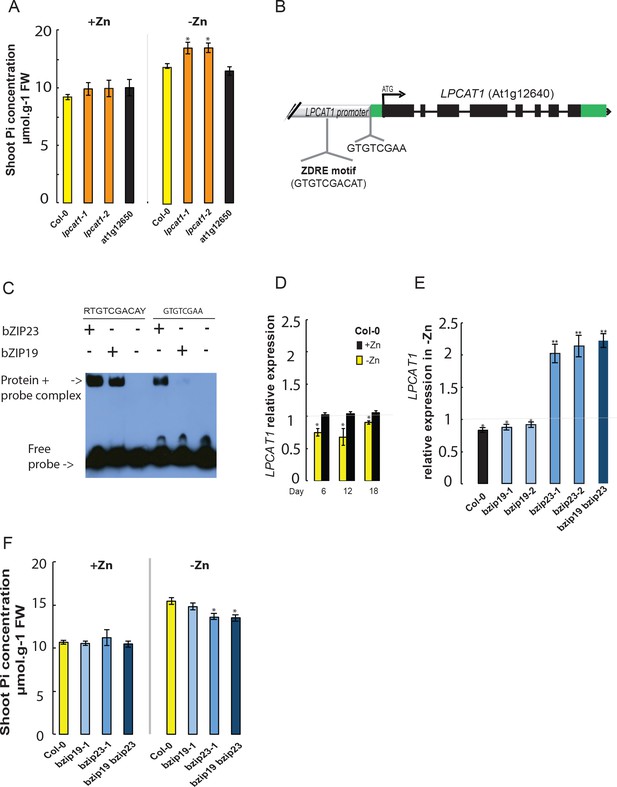
Loss of function mutation of Lyso-PhosphatidylCholine AcylTransferase 1 (LPCAT1), and not At1g12650, affects shoot Pi concentration in a Zn supply and bZIP23 dependent manner.
(A) Shoot Pi concentration of 18-days-old Col-0 wild-type plants, lpcat1 and At1g12650 mutants grown in +Zn or –Zn conditions. (B) Gene structure of LPCAT1. The grey box represents the promoter region, green boxes are 5’ and 3’ untranslated regions, black boxes represent exons, and black lines represent introns, the arrow head indicates the direction of transcription, ATG indicates the start codon. The Zinc Deficiency Response Element (ZDRE) binding site for bZIP19 and bZIP23, and the newly identified binding site for bZIP23 are indicated. (C) Differential binding of bZIP19 and bZIP23 to two promoter regions of LPCAT1 gene. EMSA analysis on 30 bp promoter fragments from motif present in LPCAT1 promoter of contrasting accessions showed in (A). (D) Relative LPCAT1 transcript abundance (-Zn/+Zn) in Col-0 wild-type plants grown on +Zn or -Zn agar medium for 6, 12 and 18 days. (E) Relative LPCAT1 transcript abundance in Col-0 wild-type plants, bzip19, bzip23, and bzip19/bzip23 double mutants grown on +Zn or -Zn agar medium for 18 days. The relative mRNA levels was quantified by RT-qPCR and normalized to the Ubiquitin10 reference mRNA level (UBQ10: At4g05320). (F) Shoot Pi concentration in Col-0 wild-type plants, bzip19 and bzip19/bzip23 double mutants grown on +Zn or -Zn agar medium for 18 days. Values are means of three to six biological replicates. Individual measurements were obtained from the analysis of shoots collected from a pool of 10 plants. Error bars indicate SD; One and two asterisks indicate a significant difference with WT plants (ANOVA and Tukey test) of p<0.05 and p<0.01, respectively.
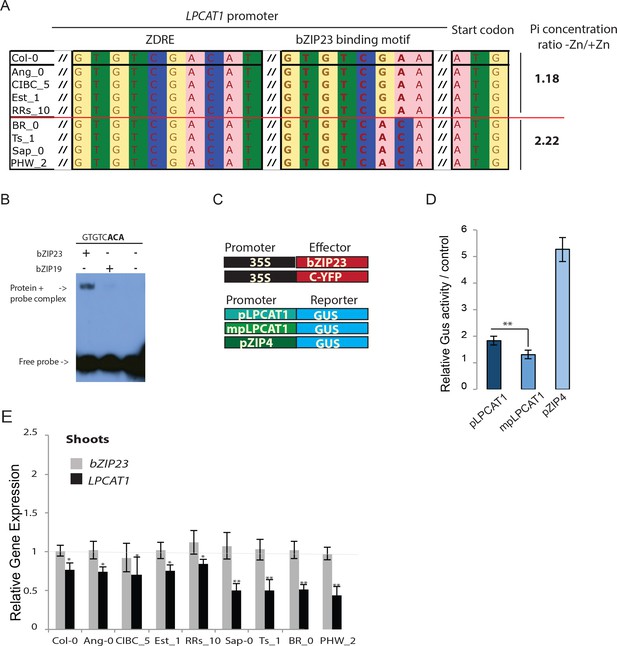
Identification of a new binding motif specific for bZIP23, and the variation of LPCAT1 gene expression between genotypes in -Zn condition.
(A) Sequence comparison for ZDRE and the new binding site motif for bZIP23 in the promoter of accession with high ratio of Pi accumulation in -Zn/+Zn (Col-0, Ang-0, CICB-5, Est-1, RRS-10) and low Pi accumulation ratio - Zn/+Zn (Sap-0, Ts-1, Br-0 and PHW-2). (B) Differential binding of bZIP19 and bZIP23 to a specific ZDRE motif of LPCAT1 (GTGTCACA). (C–D) in planta transactivation assay. (C) 35S:bZIP23 and 35S::C-YFP were used as effectors. pLPCAT1: the native Col-0 LPACT1 promoter (with « GTGTCGAA » as new ZDRE), mpLPCAT1: a modified (point mutation) version of the Col-0 promoter to only contain the new ZDRE of Sap-0 (« GTGTCACA »); pZIP4; promoter of the zinc transporter ZIP4 gene. Each pLPCAT1 (native, Col-0), mpLPCAT1 (mutated version) or pZIP4 promoter was fused to a β-glucuronidase (GUS)-encoding reporter gene (reporter). (D) The effect of bZIP23 TF on the activity of each promoter pLPCAT1, mpLPCAT1 or pZIP4 was determined by measuring GUS activity. The effect of C-YFP protein on the activity of each promoter pLPCAT1, mpLPCAT1 or pZIP4 was used as a control to determine the basal level of GUS activity for each promoter. Comparing the effect of bZIP23 TF and C-YFP protein on each promoter enabled the determination of the relative GUS activity. Error bars represent standard error from three independent experiments. The asterisks indicate that the relative GUS activity is statistically different from the YFP control (p-value<0.01, t-test). (E) Relative bZIP23 and LPCAT1 transcripts abundance in -Zn and +Zn conditions. Col-0, Ang-0, CICB-5, Est-1, RRS-10, Sap-0, Ts-1, Br-0 and PHW-2 genotypes were grown on +Zn or -Zn agar medium. The relative mRNA level was quantified by RT-qPCR and normalized to the Ubiquitin10 reference mRNA level (UBQ10: At4g05320). Values are means of six biological replicates. Individual measurements were obtained from the analysis of shoots collected from a pool of 20 plants. Error bars indicate SD; one and two asterisks indicate a significant difference with Col-0 plants (ANOVA and Tukey test) of p<0.05 and p<0.01 respectively.
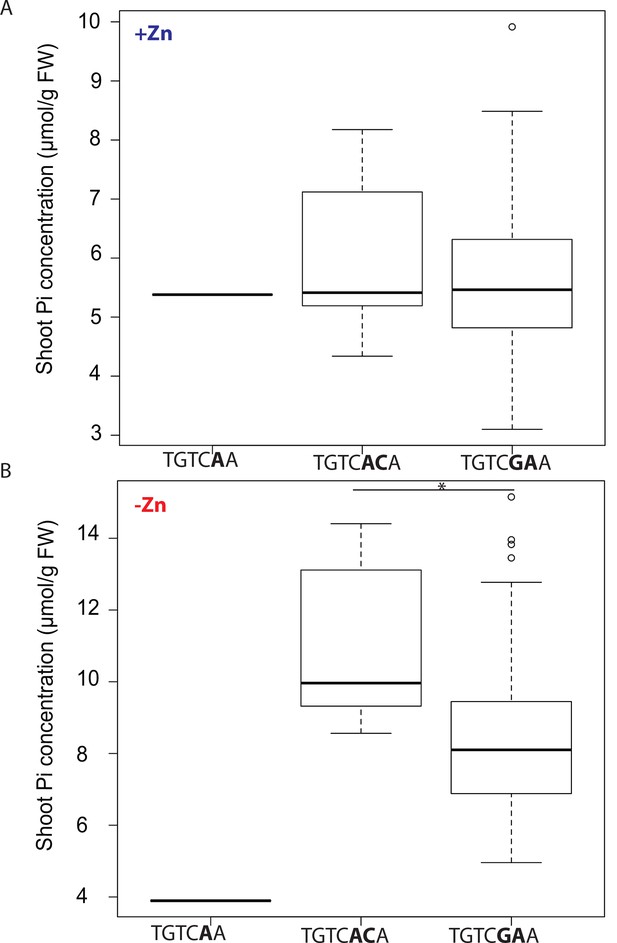
Shoot Pi concentrations in Arabidopsis accessions grouped by new ZDRE motif.
Shoot Pi concentrations of Arabidopsis thaliana accessions grown on agar medium supplemented with zinc (+Zn) (A) or without zinc (-Zn) (B) for 18 days under long day conditions. Data same as in Supplementary file 3. Grouping of accessions is based on their ZDRE binding motif allele TGTCAAA, TGTCACA and TGTCGAA respectively. Asterisk indicates a significant difference according to Student’s T-test p<0.01.
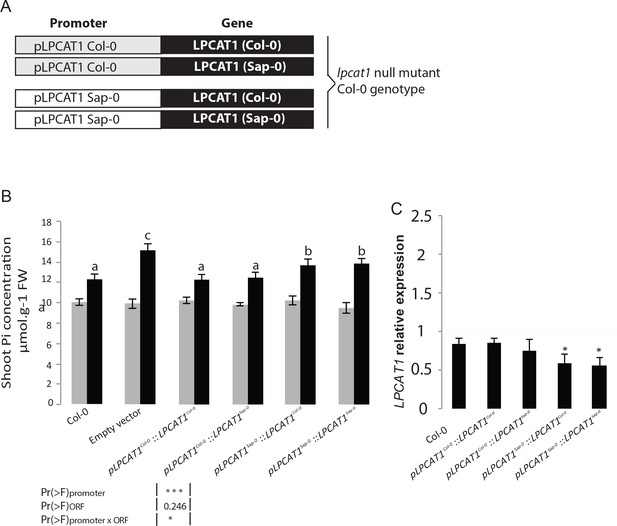
Natural allelic variation of LPCAT1 locus causes phenotypic variation of Pi accumulation in Zn deficiency conditions.
(A) Schematic representation of the transgenic constructs used to complement the lpcat1 null mutant (Col-0 background). (B) Shoot Pi concentration (–Zn /+Zn) of 18-days-old Col-0 wild-type plants, lpcat1 mutant transformed with empty vector, or with constructs schematized in (A) grown in +Zn or –Zn conditions. (C) The ANOVA results are presented in the table. Significative codes: ‘***’ 0.001 and ‘*’ 0.05. Relative LPCAT1 transcript abundance in wild-type plants (Col-0 background) and the transgenic lines generated using the construct schematized in (A) grown on +Zn or -Zn agar medium. The relative mRNA levels was quantified by RT-qPCR and normalized to the Ubiquitin10 reference mRNA level (UBQ10: At4g05320). Values are means of three to biological replicates. Individual measurements were obtained from the analysis of shoots collected from a pool of six plants. Error bars indicate SD; asterisks indicates a significant difference with Col-0 plants (ANOVA and Tukey test) of p<0.05.
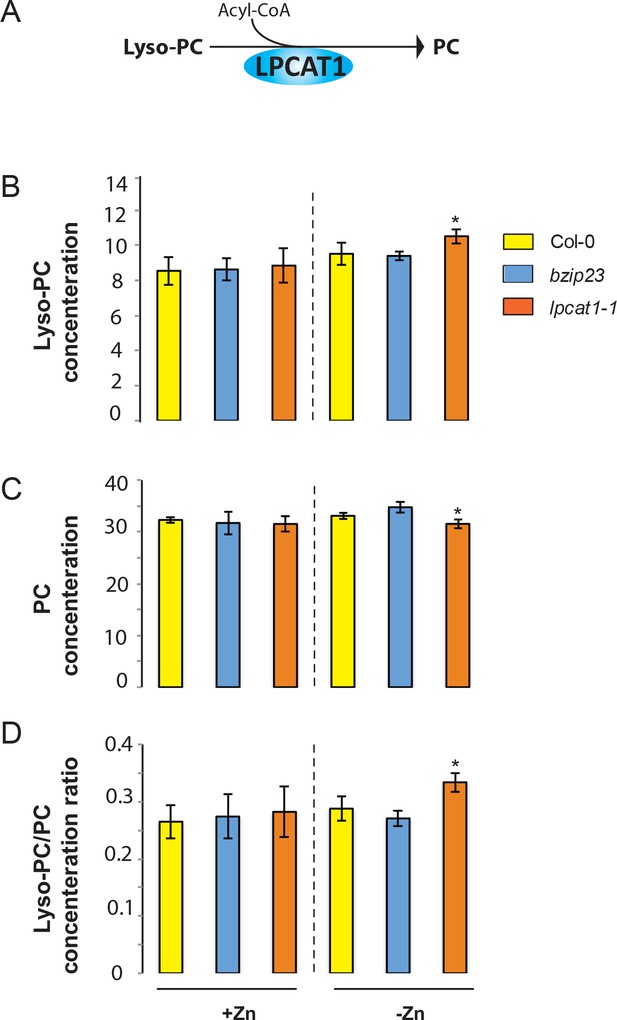
Loss of function mutations of LPCAT1 affects the lysoPC/PC ratio in –Zn conditions.
(A) Schematic representation of the biochemical function of LPCAT1, which catalyses the formation of phosphatidylcholine (PC) from lyso-PC and long-chain acyl-CoA. (B) Lyso-PC concentration (C) PC concentration (D) Lyso-PC/PC concentration ratios of Col-0 wild-type plants, bzip23 and lpcat1 mutant lines grown in +Zn or -Zn conditions for 18 days. Individual measurements were obtained from the analysis of shoots collected from a pool of five plants. Data are mean ±SD of three biological replicates. Statistically significant differences (ANOVA and Tukey test, p<0.05) between mutants and Col-0 are indicated with asterisks.
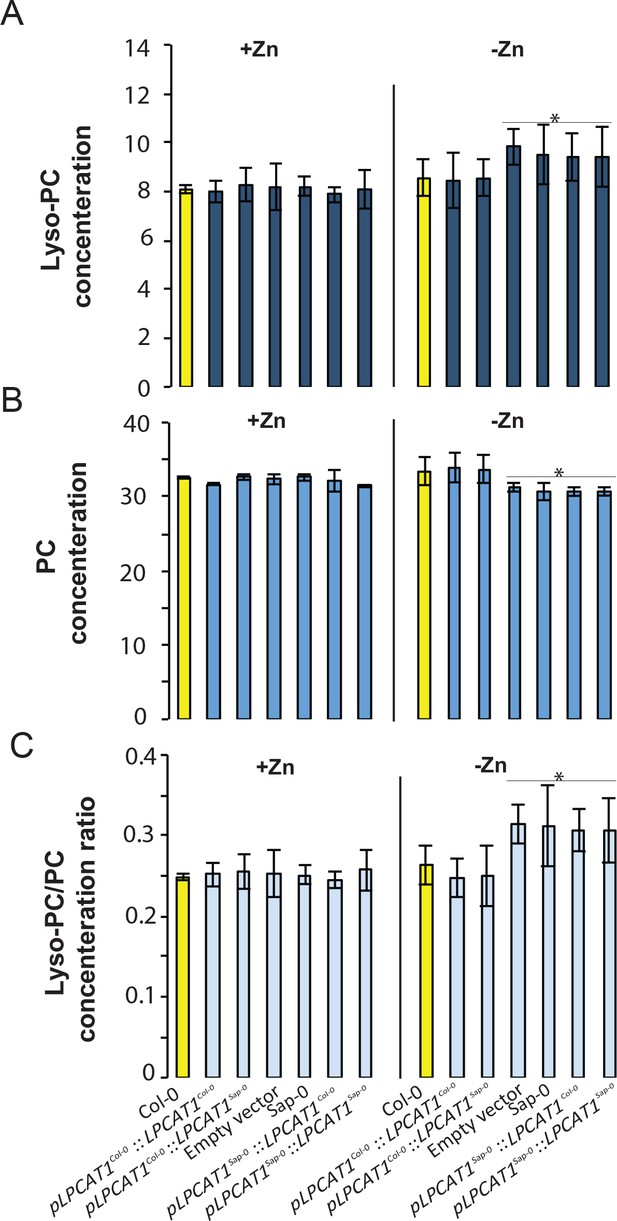
Effect of the polymorphisms in the regulatory region of LPCAT1 on the change in LPC/PC ratios in –Zn conditions.
(A) Lyso-PC concentration (B) PC concentration (C) Lyso-PC/PC concentration ratios of Sap-0, Col-0 wild-type plants, and lpcat1 expressing pLPCAT1Col-0::LPCAT1Col-0, pLPCAT1Col-0::LPCAT1Sap-0, pLPCAT1Sap-0 ::LPCAT1Col-0, pLPCAT1Sap-0::LPCAT1Sap-0 constructs and lpcat1 transformed with empty lines grown in +Zn or -Zn conditions for 18 days. Individual measurements were obtained from the analysis of shoots collected from a pool of five plants. Data are mean ±SD of three biological replicates. Statistically significant differences (ANOVA and Tukey test, p<0.05) between mutants and Col-0 are indicated with asterisks.
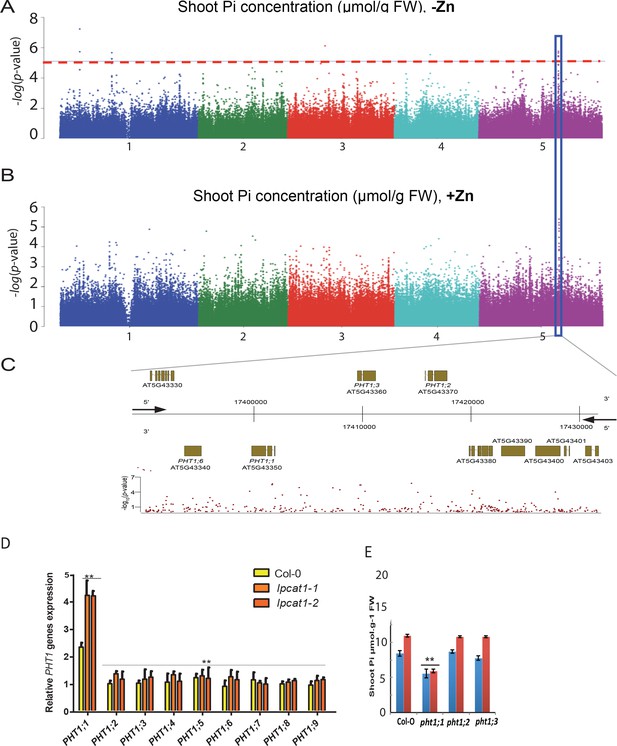
Loss of function mutations of LPCAT1 show enhanced expression of PHT1;1 when compared to Col-0 wild-type plants.
(A, B) Genome-wide association (GWA) analysis of Arabidopsis shoot Pi concentration. 223 Arabidopsis thaliana accessions were grown on agar medium supplemented with zinc (+Zn) or without zinc (-Zn) for 18 days under long day conditions, upon which shoot inorganic phosphate (Pi) concentrations were determined. Manhattan plots of GWA analysis of Arabidopsis shoot Pi concentration in -Zn (A) and +Zn (B). The five Arabidopsis chromosomes are indicated in different colours. Each dot represents the –log10(P) association score of one single nucleotide polymorphism (SNP). The dashed red line denotes an approximate false discovery rate 10% threshold. Boxes indicate the location of the PHT1 (blue) quantitative trait loci (QTL). (C) Gene models (upper panel) and SNP –log10(P) scores (lower panel) in the genomic region surrounding the GWA QTL at the or PHT1 locus; 5’ and 3’ indicate the different genomic DNA strands and orientation of the respective gene models. (D) Relative expression level of all members of the Arabidopsis PHT1 gene family in shoots of 18-days-old Col-0 wild-type plants and lpcat1 mutants grown on -Zn agar medium compared to their expression on +Zn. mRNA accumulation was quantified by RT-qPCR, normalized to the mRNA level of the UBIQUITIN10 reference gene (UBQ10: At4g05320) and expressed as relative values against its UBQ10 normalized mRNA level of Col-0 grown in +Zn medium (control). (E) Shoot Pi concentration of 18-days-old Col-0 wild-type plants, pht1;1, pht1;2, and pht1;3 mutants grown in +Zn or –Zn conditions. Data are mean ±SD of three biological replicates. Statistically significant differences (ANOVA and Tukey test, p<0.05 and p<0.01) are indicated by one or two asterisks.
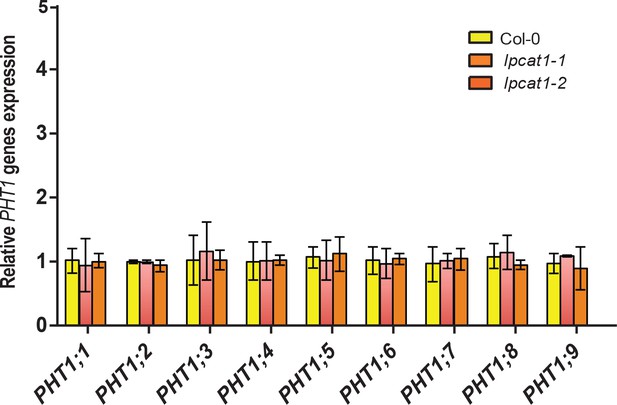
Relative expression level of all members of the Arabidopsis PHT1 gene family.
Arabidopsis thaliana Col-0, lpcat1-1 and lpcat1-2 mutant lines were grown in agar medium supplemented with zinc (+Zn) for 18 days under long day conditions. mRNA level was quantified by RT-qPCR and normalized to the Ubiquitin10 reference mRNA level (UBQ10: At4g05320) and expressed as relative values against the UBQ10 normalized mRNA level in Col-0. Values are means of six biological replicates. Individual measurements were obtained from the analysis of shoots collected from a pool of 20 plants. Error bars indicate SD; one and two asterisk indicates a significant difference with Col-0 plants (ANOVA and Tukey test) of p<0.05 and p<0.01 respectively.
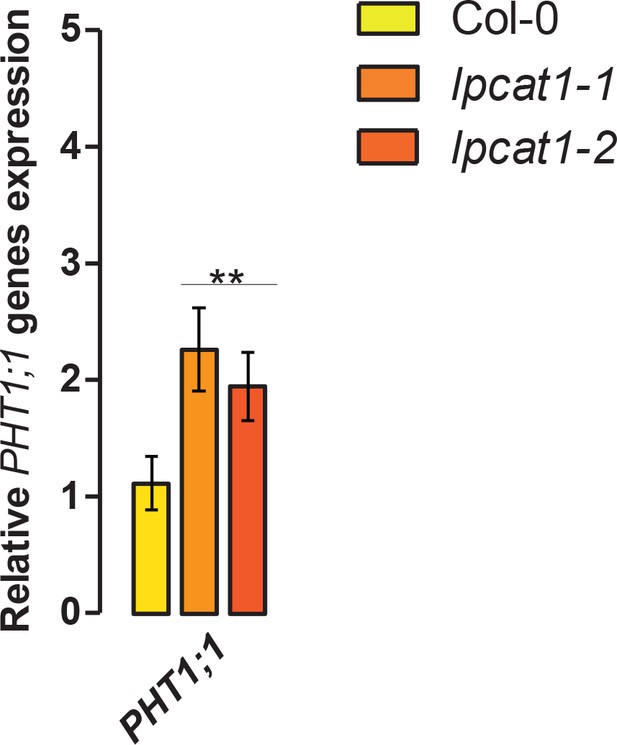
High Affinity of Phosphate Transporter (PHT1;1) gene expression analysis.
Loss of function mutations of LPCAT1 show enhanced expression of PHT1;1 when compared to Col-0 wild-type plants. Relative expression level of PHT1;1 gene in roots of 18-days-old Col-0 wild-type plants and lpcat1 mutants grown on -Zn agar medium. mRNA accumulation was quantified by RT-qPCR, normalized to the mRNA level of the UBIQUITIN10 reference gene (UBQ10: At4g05320) and expressed as relative values against Col-0 grown in +Zn medium (control). Individual measurements were obtained from the analysis of roots collected from a pool of five plants. Data are mean ±SD of three biological replicates. Statistically significant differences (ANOVA and Tukey test, p<0.01) are indicated by two asterisks.
Additional files
-
Supplementary file 1
Shoots Pi concentration in the 223 Arabidopsis thaliana accessions grown in presence or absence of zinc.
- https://doi.org/10.7554/eLife.32077.015
-
Supplementary file 2
Coordinates for the significant SNPs associated with Pi concentration in shoots in Zn conditions.
- https://doi.org/10.7554/eLife.32077.016
-
Supplementary file 3
List of new ZDRE motif in the Arabidopsis thaliana accessions and the shoot Pi content in presence (+Zn) or absence of Zn (-Zn).
- https://doi.org/10.7554/eLife.32077.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32077.018





