Neutrophil-generated HOCl leads to non-specific thiol oxidation in phagocytized bacteria
Figures
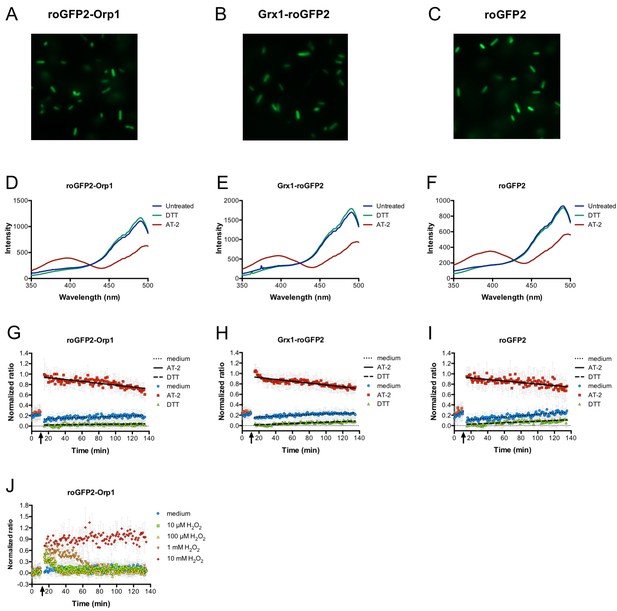
Expression of genetically encoded redox probes in E. coli.
Fluorescence microscopy reveals uniform expression of roGFP2-based probes (fused to Orp1 and Grx1 and unfused probe) from a plasmid in the cytoplasm of E. coli (A–C). The probes’ response to the strong thiol reductant DTT and the strong thiol oxidant AT-2 could be measured in a fluorescence spectrophotometer by monitoring the characteristic excitation spectra (D–F). A normalized ratio of the intensity of fluorescence at 405 and 488 nm excitation allowed for a time-course measurement of the probes’ oxidation in response to DTT and AT-2 treatment in a 96-well plate reader. The arrows indicate the time point of addition of the oxidant and reductant. Medium served as a control (G–I). The level of oxidation caused by hydrogen peroxide in the hydrogen peroxide-sensitive probe roGFP2-Orp1 expressed in E. coli can be followed in a plate reader. The arrow indicates the addition of hydrogen peroxide at the concentrations indicated (J).
-
Figure 1—source data 1
Numerical fluorescence spectrometry data represented in Figure 1D.
- https://doi.org/10.7554/eLife.32288.004
-
Figure 1—source data 2
Numerical fluorescence spectrometry data represented in Figure 1E.
- https://doi.org/10.7554/eLife.32288.005
-
Figure 1—source data 3
Numerical fluorescence spectrometry data represented in Figure 1F.
- https://doi.org/10.7554/eLife.32288.006
-
Figure 1—source data 4
Numerical fluorescence plate reader data represented in Figure 1G.
- https://doi.org/10.7554/eLife.32288.007
-
Figure 1—source data 5
Numerical fluorescence plate reader data represented in Figure 1H.
- https://doi.org/10.7554/eLife.32288.008
-
Figure 1—source data 6
Numerical fluorescence plate reader data represented in Figure 1I.
- https://doi.org/10.7554/eLife.32288.009
-
Figure 1—source data 7
Numerical fluorescence plate reader data represented in Figure 1J.
- https://doi.org/10.7554/eLife.32288.010
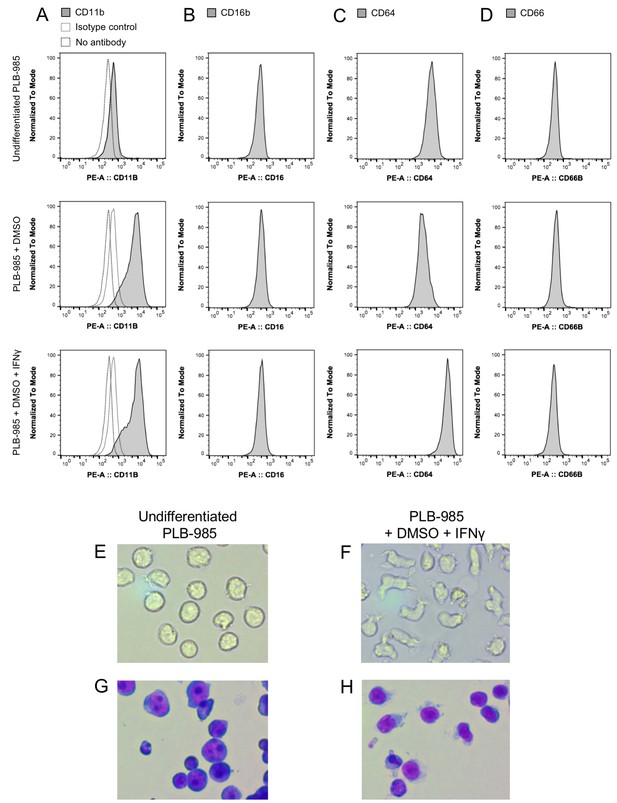
Differentiation of the myeloid cell line PLB-985 to neutrophil-like cells.
Determination of surface markers CD11b, CD16b, CD64, and CD66 before and after two different differentiation protocols (A–D). Cells differentiated with DMSO or DMSO and IFNγ expressed significantly higher levels of the neutrophil markers CD11b and CD64 when compared to undifferentiated cells. These changes in the expression of surface markers coincided with morphological changes typical for neutrophils when observed under phase contrast (E–F) or stained with May-Grünwald-Giemsa stain (G–H).
-
Figure 2—source data 1
Numerical flow cytometry data represented in Figure 2A, trace CD11b, Undifferentiated.
- https://doi.org/10.7554/eLife.32288.012
-
Figure 2—source data 2
Numerical flow cytometry data represented in Figure 2A, trace CD11b, Undifferentiated, isotype control.
- https://doi.org/10.7554/eLife.32288.013
-
Figure 2—source data 3
Numerical flow cytometry data represented in Figure 2A, trace CD11b, Undifferentiated, negative control.
- https://doi.org/10.7554/eLife.32288.014
-
Figure 2—source data 4
Numerical flow cytometry data represented in Figure 2A, trace CD11b, DMSO.
- https://doi.org/10.7554/eLife.32288.015
-
Figure 2—source data 5
Numerical flow cytometry data represented in Figure 2A, trace CD11b, DMSO, isotype control.
- https://doi.org/10.7554/eLife.32288.016
-
Figure 2—source data 6
Numerical flow cytometry data represented in Figure 2A, trace CD11b, DMSO, negative control.
- https://doi.org/10.7554/eLife.32288.017
-
Figure 2—source data 7
Numerical flow cytometry data represented in Figure 2A, trace CD11b, DMSO+ IFNγ.
- https://doi.org/10.7554/eLife.32288.018
-
Figure 2—source data 8
Numerical flow cytometry data represented in Figure 2A, trace CD11b, DMSO+ IFNγ, isotype control.
- https://doi.org/10.7554/eLife.32288.019
-
Figure 2—source data 9
Numerical flow cytometry data represented in Figure 2A, trace CD11b, DMSO+ IFNγ, negative control.
- https://doi.org/10.7554/eLife.32288.020
-
Figure 2—source data 10
Numerical flow cytometry data represented in Figure 2B, trace CD16, Undifferentiated.
- https://doi.org/10.7554/eLife.32288.021
-
Figure 2—source data 11
Numerical flow cytometry data represented in Figure 2B, trace CD16, DMSO.
- https://doi.org/10.7554/eLife.32288.022
-
Figure 2—source data 12
Numerical flow cytometry data represented in Figure 2B, trace CD16, DMSO+ IFNγ.
- https://doi.org/10.7554/eLife.32288.023
-
Figure 2—source data 13
Numerical flow cytometry data represented in Figure 2C, trace CD64, Undifferentiated.
- https://doi.org/10.7554/eLife.32288.024
-
Figure 2—source data 14
Numerical flow cytometry data represented in Figure 2C, trace CD64, DMSO.
- https://doi.org/10.7554/eLife.32288.025
-
Figure 2—source data 15
Numerical flow cytometry data represented in Figure 2C, trace CD64, DMSO+ IFNγ.
- https://doi.org/10.7554/eLife.32288.026
-
Figure 2—source data 16
Numerical flow cytometry data represented in Figure 2D, trace CD66b, Undifferentiated.
- https://doi.org/10.7554/eLife.32288.027
-
Figure 2—source data 17
Numerical flow cytometry data represented in Figure 2D, trace CD66b, DMSO.
- https://doi.org/10.7554/eLife.32288.028
-
Figure 2—source data 18
Numerical flow cytometry data represented in Figure 2D, trace CD66b, DMSO+ IFNγ.
- https://doi.org/10.7554/eLife.32288.029
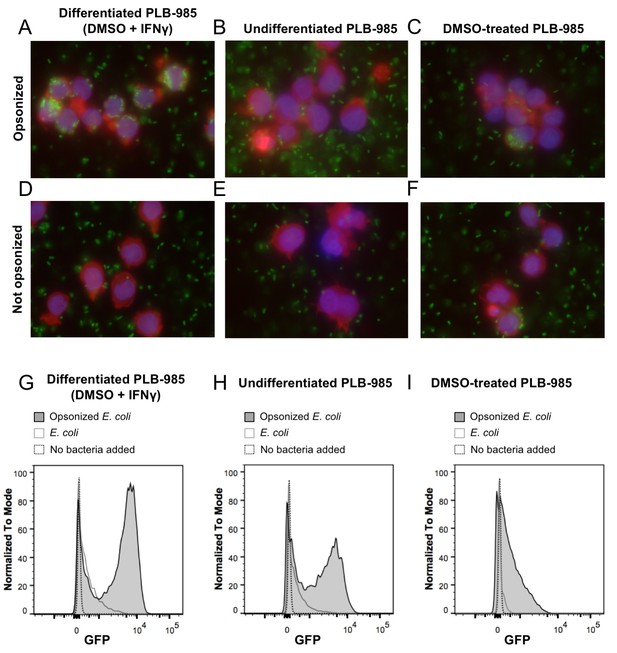
Phagocytosis of E. coli by PLB-985.
(A) PLB-985 cells differentiated with DMSO and IFNγ effectively phagocytize E. coli opsonized with human immunoglobulin G (IgG). E. coli cells expressing roGFP2-Orp1 associate with the neutrophil-like cells. This is not the case with undifferentiated cells (B). PLB-985 cells treated with DMSO are less effective in their phagocytosis as well (C). Effective phagocytosis is dependent on opsonization (D–F). Microscopic images are composite overlays of the DAPI nuclear stain (blue), the TRITC-conjugated phalloidin-based actin stain (red) and the GFP channel (bacteria, green). Flow cytometry corroborates the microscopic evidence. Differentiated cells co-incubated with opsonized E. coli show the highest population of cells incorporating bacteria as measured by GFP-fluorescence (G), whereas the bacteria-incorporating population size in undifferentiated (H) and DMSO-treated cells (I) is significantly smaller.
-
Figure 3—source data 1
Numerical flow cytometry data represented in Figure 3G, trace Opsonized E. coli.
- https://doi.org/10.7554/eLife.32288.031
-
Figure 3—source data 2
Numerical flow cytometry data represented in Figure 3G, trace E. coli.
- https://doi.org/10.7554/eLife.32288.032
-
Figure 3—source data 3
Numerical flow cytometry data represented in Figure 3G, trace No bacteria added.
- https://doi.org/10.7554/eLife.32288.033
-
Figure 3—source data 4
Numerical flow cytometry data represented in Figure 3H, trace Opsonized E. coli.
- https://doi.org/10.7554/eLife.32288.034
-
Figure 3—source data 5
Numerical flow cytometry data represented in Figure 3H, trace E. coli.
- https://doi.org/10.7554/eLife.32288.035
-
Figure 3—source data 6
Numerical flow cytometry data represented in Figure 3H, trace no bacteria added.
- https://doi.org/10.7554/eLife.32288.036
-
Figure 3—source data 7
Numerical flow cytometry data represented in Figure 3I, trace Opsonized E. coli.
- https://doi.org/10.7554/eLife.32288.037
-
Figure 3—source data 8
Numerical flow cytometry data represented in Figure 3I, trace E. coli.
- https://doi.org/10.7554/eLife.32288.038
-
Figure 3—source data 9
Numerical flow cytometry data represented in Figure 3I, trace no bacteria added.
- https://doi.org/10.7554/eLife.32288.039
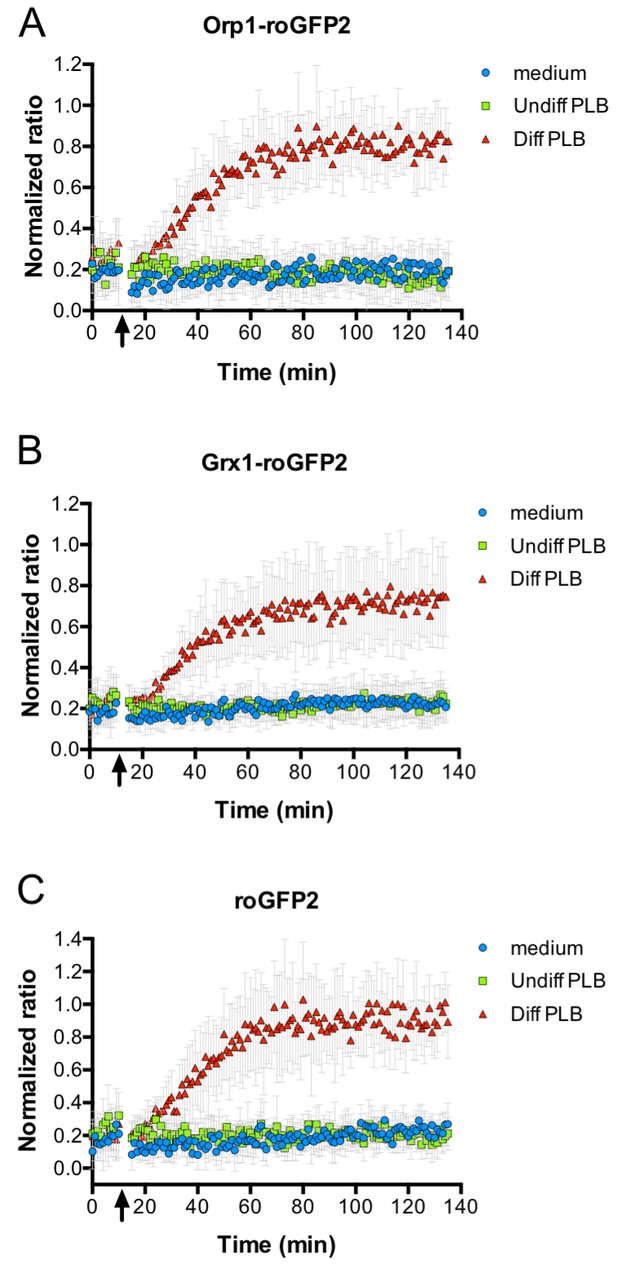
Oxidation of roGFP2-based probes expressed in E. coli co-cultivated with neutrophil-like cells.
E. coli cells expressing roGFP2-Orp1 from a plasmid were incubated in a 96-well plate reader. The ratio of the fluorescence intensity at excitation wavelengths of 405 and 488 nm was calculated and plotted over time. The arrow indicates the addition of medium, undifferentiated PLB-985 cells and differentiated, neutrophil-like PLB-985. Addition of neutrophil-like cells led to probe oxidation, as evidenced by the increase in the ratio of the fluorescence intensities (A). The probe oxidation in bacteria expressing the Grx1-roGFP2 fusion probe (B) and unfused roGFP2 (C) showed kinetics virtually identical to E. coli cells expressing roGFP2-Orp1.
-
Figure 4—source data 1
Numerical fluorescence plate reader data represented in Figure 4A.
- https://doi.org/10.7554/eLife.32288.041
-
Figure 4—source data 2
Numerical fluorescence plate reader data represented in Figure 4B.
- https://doi.org/10.7554/eLife.32288.042
-
Figure 4—source data 3
Numerical fluorescence plate reader data represented in Figure 4C.
- https://doi.org/10.7554/eLife.32288.043
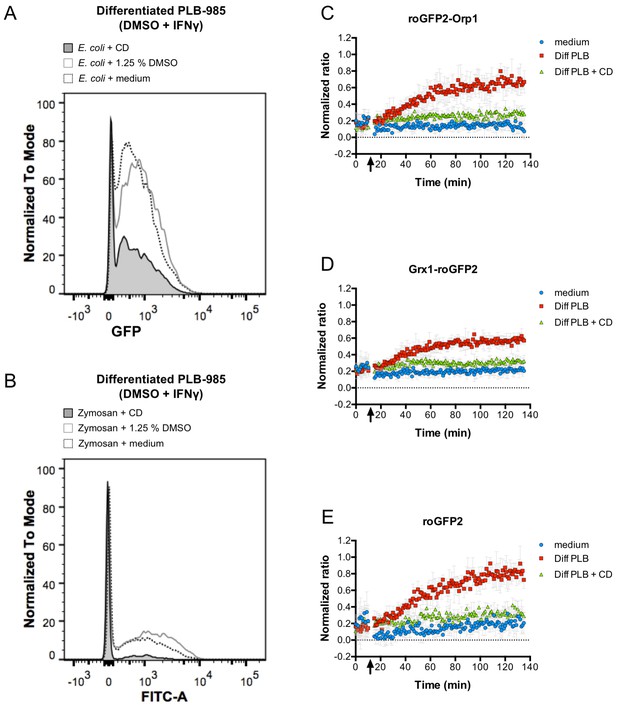
Phagocytosis is required for efficient oxidation of roGFP2-based probes.
Cytochalasin D inhibits phagocytosis of E. coli (A) and FITC-labeled Zymosan (B) as determined by flow cytometry. Cytochalasin D (CD) treatment of differentiated PLB-985 cells inhibits oxidation of roGFP2-Orp1 expressed in E. coli (C). Similar findings were observed in bacteria expressing Grx1-roGFP2 (D) and unfused roGFP2 (E). Arrows indicate the addition of medium, differentiated, neutrophil-like PLB-985 cells and cytochalasin D-treated differentiated PLB-985 cells.
-
Figure 5—source data 1
Numerical flow cytometry data represented in Figure 5A, trace E. coli + CD.
- https://doi.org/10.7554/eLife.32288.045
-
Figure 5—source data 2
Numerical flow cytometry data represented in Figure 5A, trace E. coli + 1.25% DMSO.
- https://doi.org/10.7554/eLife.32288.046
-
Figure 5—source data 3
Numerical flow cytometry data represented in Figure 5A, trace E. coli + medium.
- https://doi.org/10.7554/eLife.32288.047
-
Figure 5—source data 4
Numerical flow cytometry data represented in Figure 5B, trace Zymosan + CD.
- https://doi.org/10.7554/eLife.32288.048
-
Figure 5—source data 5
Numerical flow cytometry data represented in Figure 5B, trace Zymosan + 1.25% DMSO.
- https://doi.org/10.7554/eLife.32288.049
-
Figure 5—source data 6
Numerical flow cytometry data represented in Figure 5B, trace Zymosan + medium.
- https://doi.org/10.7554/eLife.32288.050
-
Figure 5—source data 7
Numerical fluorescence plate reader data represented in Figure 5C.
- https://doi.org/10.7554/eLife.32288.051
-
Figure 5—source data 8
Numerical fluorescence plate reader data represented in Figure 5D.
- https://doi.org/10.7554/eLife.32288.052
-
Figure 5—source data 9
Numerical fluorescence plate reader data represented in Figure 5E.
- https://doi.org/10.7554/eLife.32288.053
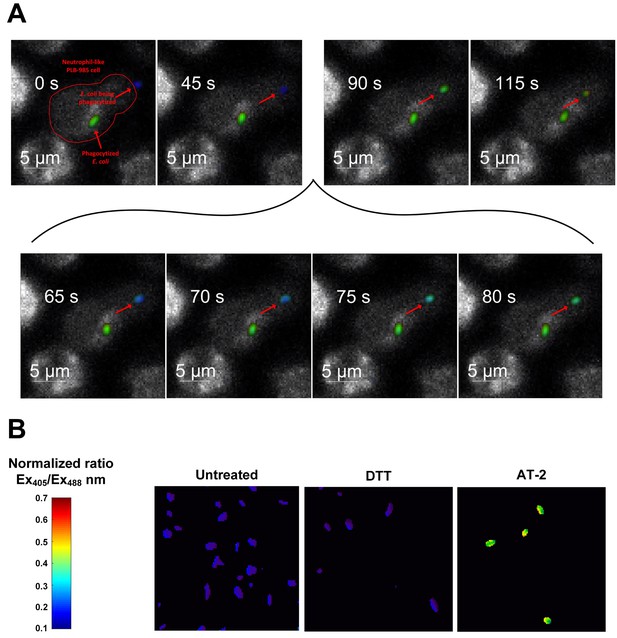
Probe oxidation occurs within seconds after phagocytosis.
(A) Quantitative fluorescence microscopy of the redox state of roGFP2-Orp1 in E. coli during phagocytosis. Stills of a movie observing an individual E. coli cell expressing roGFP2-Orp1 (indicated by an arrow) being attacked by a neutrophil-like PLB-985 cell. The neutrophil has already phagocytized another E. coli cell. Upon phagocytosis, the oxidation state changes within seconds (inset 65–80 s) as illustrated based on the false color scale indicated. See also Video 1. (B) Control: E. coli cells in the absence of neutrophil-like cells untreated and treated with the reducing agent dithiothreitol (DTT) and the oxidizing agent Aldrithiol-2 (AT-2).
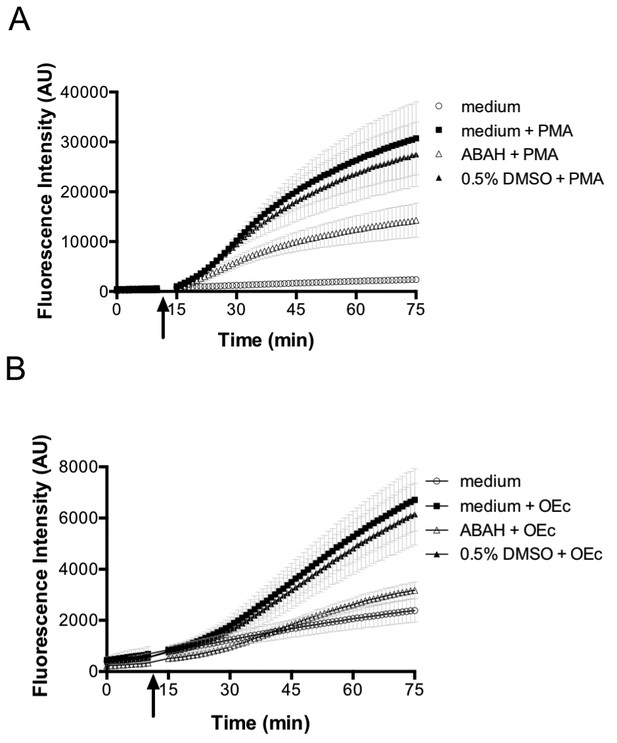
PMA and opsonized E. coli stimulate ROS production in differentiated neutrophil-like PLB-985 cells.
Differentiated, neutrophil-like PLB-985 produce reactive species that can be detected by 2',7'-Dichlordihydrofluorescein-diacetate (H2DCFDA) oxidation when stimulated with PMA (A) or opsonized E. coli (B). The myeloperoxidase inhibitor ABAH (4-aminobenzoic acid hydrazide) decreases the production of reactive species. The arrow indicates the time-point of the addition of PMA or opsonized E. coli.
-
Figure 7—source data 1
Numerical fluorescence plate reader data represented in Figure 7.
- https://doi.org/10.7554/eLife.32288.057
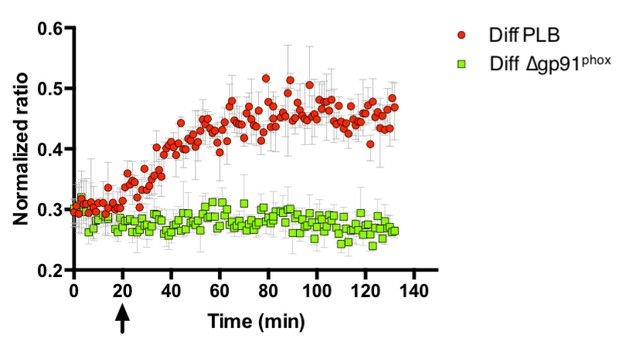
Probe oxidation in bacteria is dependent on NOX2.
PLB-985 lacking gp91phox, the catalytic subunit of NOX2 and a major intracellular producer of superoxide, no longer induce probe oxidation in E. coli cells expressing roGFP2-Orp1. The arrow indicates the addition of differentiated, neutrophil-like PLB-985 cells or differentiated PLB-985 cells lacking gp91phox.
-
Figure 8—source data 1
Numerical fluorescence plate reader data represented in Figure 8.
- https://doi.org/10.7554/eLife.32288.059
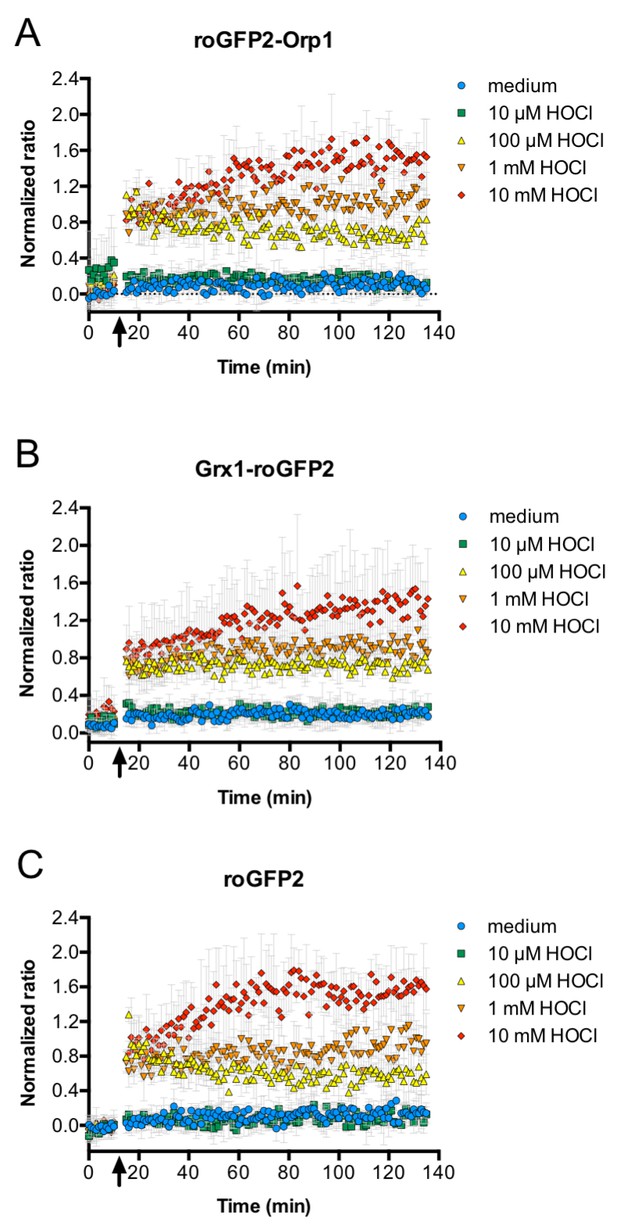
HOCl oxidizes roGFP2-based probes in E. coli effectively and with fast kinetics.
All three roGFP2-based probes used in this study, roGFP2-Orp1 (A), Grx1-roGFP2 (B), and unfused roGFP2(C) are oxidized within mixing time upon addition of HOCl. E. coli cells expressing the respective probes were incubated in a 96-well plate reader and a time-course measurement of the ratio of the fluorescence intensity of the probe at excitation wavelengths of 405 and 488 nm was performed. The arrow indicates the addition of different concentrations of HOCl or medium as a control as indicated.
-
Figure 9—source data 1
Numerical fluorescence plate reader data represented in Figure 9A.
- https://doi.org/10.7554/eLife.32288.061
-
Figure 9—source data 2
Numerical fluorescence plate reader data represented in Figure 9B.
- https://doi.org/10.7554/eLife.32288.062
-
Figure 9—source data 3
Numerical fluorescence plate reader data represented in Figure 9C.
- https://doi.org/10.7554/eLife.32288.063
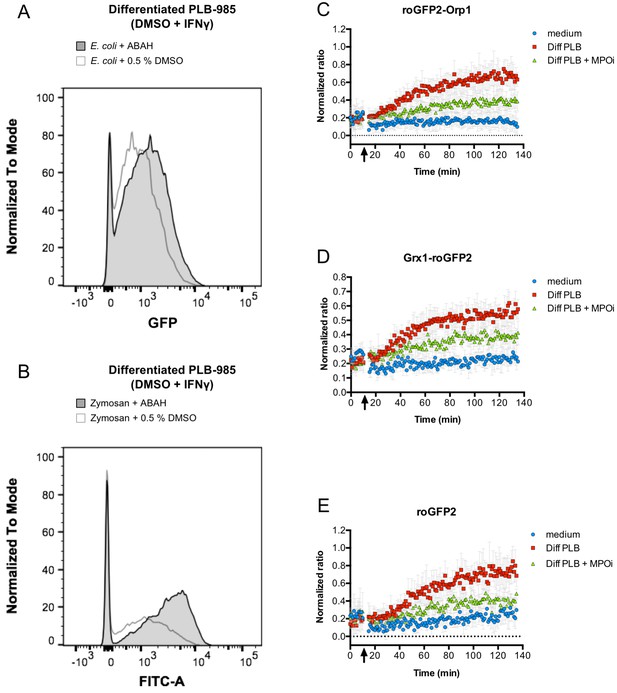
Inhibition of myeloperoxidase abrogates probe oxidation.
Addition of myeloperoxidase inhibitor 4-aminobenzoic acid hydrazide (ABAH) does not inhibit phagocytosis of opsonized E. coli (A) or Zymosan (B), as evidenced by flow cytometry. However, neutrophil-like cells pre-incubated with ABAH are less capable of oxidizing roGFP2-Orp1 (C), Grx1-roGFP2 (D), or unfused roGFP2 expressed in E. coli (E). Arrows indicate the addition of PLB 985 cells or medium to bacteria.
-
Figure 10—source data 1
Numerical flow cytometry data represented in Figure 10A, trace E. coli + ABAH.
- https://doi.org/10.7554/eLife.32288.065
-
Figure 10—source data 2
Numerical flow cytometry data represented in Figure 10A, trace E. coli + 0.5% DMSO.
- https://doi.org/10.7554/eLife.32288.066
-
Figure 10—source data 3
Numerical flow cytometry data represented in Figure 10A, trace Zymosan + ABAH.
- https://doi.org/10.7554/eLife.32288.067
-
Figure 10—source data 4
Numerical flow cytometry data represented in Figure 10A, trace Zymosan + 0.5% DMSO.
- https://doi.org/10.7554/eLife.32288.068
-
Figure 10—source data 5
Numerical fluorescence plate reader data represented in Figure 10C.
- https://doi.org/10.7554/eLife.32288.069
-
Figure 10—source data 6
Numerical fluorescence plate reader data represented in Figure 10D.
- https://doi.org/10.7554/eLife.32288.070
-
Figure 10—source data 7
Numerical fluorescence plate reader data represented in Figure 10E.
- https://doi.org/10.7554/eLife.32288.071
Videos
Time lapsed movie of quantitative fluorescence microscopy of the redox state of roGFP2-Orp1 in E. coli during phagocytosis.
Related to Figure 6. An E. coli cell is being attacked by a neutrophil-like PLB-985 cell. The neutrophil has already phagocytized another E. coli cell. Upon phagocytosis, the oxidation state changes within seconds.
Tables
Primers.
https://doi.org/10.7554/eLife.32288.072| Primer name | Sequence |
|---|---|
| Orp1-Fw | ccccccatatggtgagcaagggcgagga |
| Orp1-Rv | ggggggaattcttattccacctctttcaa |
| Grx-Fw | ccccccatatggctcaagagtttgtgaac |
| Grx-Rv | ggggggaattcttacttgtacagctcgtc |
Bacterial strains and plasmids.
https://doi.org/10.7554/eLife.32288.073| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1 blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac | Stratagene |
| E. coli MG1655 | F- lambda- ilvG- rfb50 rph-1 | (Blattner et al., 1997) |
| E. coli AM39 | MG1655 pCC_roGFP2-orp1 | This work |
| E. coli AM180 | MG1655 pCC_grx1-roGFP2 | This work |
| E. coli AM181 | MG1655 pCC_roGFP2 | This work |
| Plasmids | ||
| pCC | TAC-MAT-Tag-2 derivative; ptac | (Masuch et al., 2015) |
| pQE60_roGFP2-orp1-his-QC | pQE60 carrying roGFP2-orp1-his6; removed EcoRI site; pT5-lac promoter | (Müller et al., 2017a) |
| pQE60_grx1_roGFP2-his-QC | pQE60 carrying grx1-roGFP2-his6: removed EcoRI site; pT5-lac | (Müller et al., 2017a) |
| pCC_roGFP2 | roGFP2; ptac | (Müller et al., 2017a) |
| pCC_roGFP2-orp1 | roGFP2-orp1; ptac | This work |
| pCC_grx1-roGFP2 | grx1-roGFP2; ptac | This work |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32288.074





