CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo
Figures
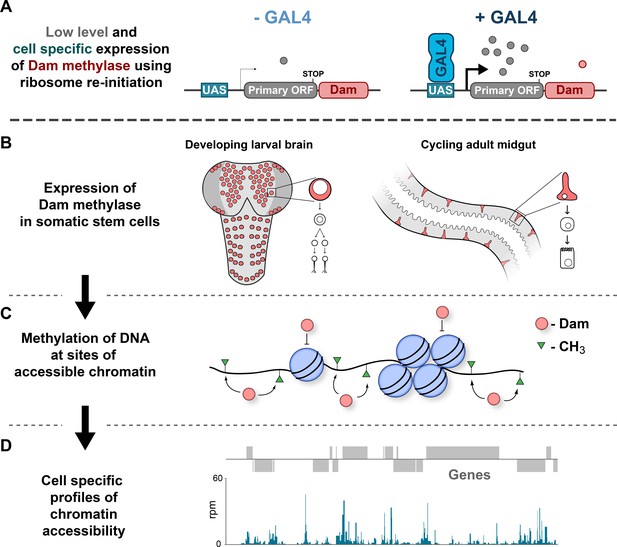
Schematic illustrating CATaDa technique.
(A–B) E. coli Dam is expressed specifically in cell-types of interest using TaDa technique. (C) GATC motifs in regions of accessible chromatin are methylated by Dam, whilst areas of condensed chromatin prevent access to Dam thereby precluding methylation. (D) Methylated DNA is detected to produce chromatin accessibility profiles for individual cell-types of interest from a mixed population of cells.
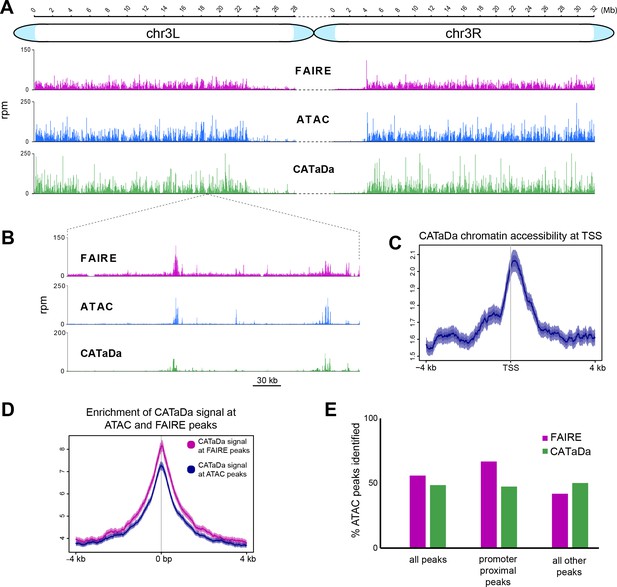
Validation of Dam chromatin accessibility profiling compared to ATAC and FAIRE-seq.
(A) Chromatin accessibility across chromosome three as determined by ATAC-seq, FAIRE-seq, and CATaDa. Note the reduced amount open chromatin proximal to the centromere regions in all three datasets. y-axes = reads per million (rpm). (B) Example locus showing data obtained by FAIRE, ATAC, and CATaDa. Peaks are broadly reproducible across techniques. (C) Aggregation plot of CATaDa signal at TSS with 2 kb regions up and downstream. Aggregated signal at TSS shows expected enrichment of Dam. (D) Aggregation plot of CATaDa signal at ATAC or FAIRE peaks, indicating enrichment of CATaDa signal at these loci. (E) Identification of ATAC peaks in CATaDa or FAIRE data. CATaDa and FAIRE identify 48.6% and 55.9% of ATAC peaks, respectively. FAIRE-seq peaks overlap more frequently with promoter proximal peaks (2 kb from TSS), whilst CATaDa peaks overlap with more ATAC peaks outside of promoter regions.
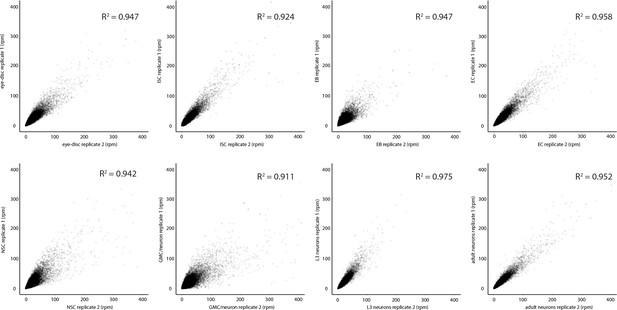
Correlation between CATaDa replicates.
Correlation between CATaDa replicates for all cell types assayed in this study shows good agreement, indicating that the technique has high reproducibility. p<2.2 × 10−16 for all replicates, indicating highly significant correlations.
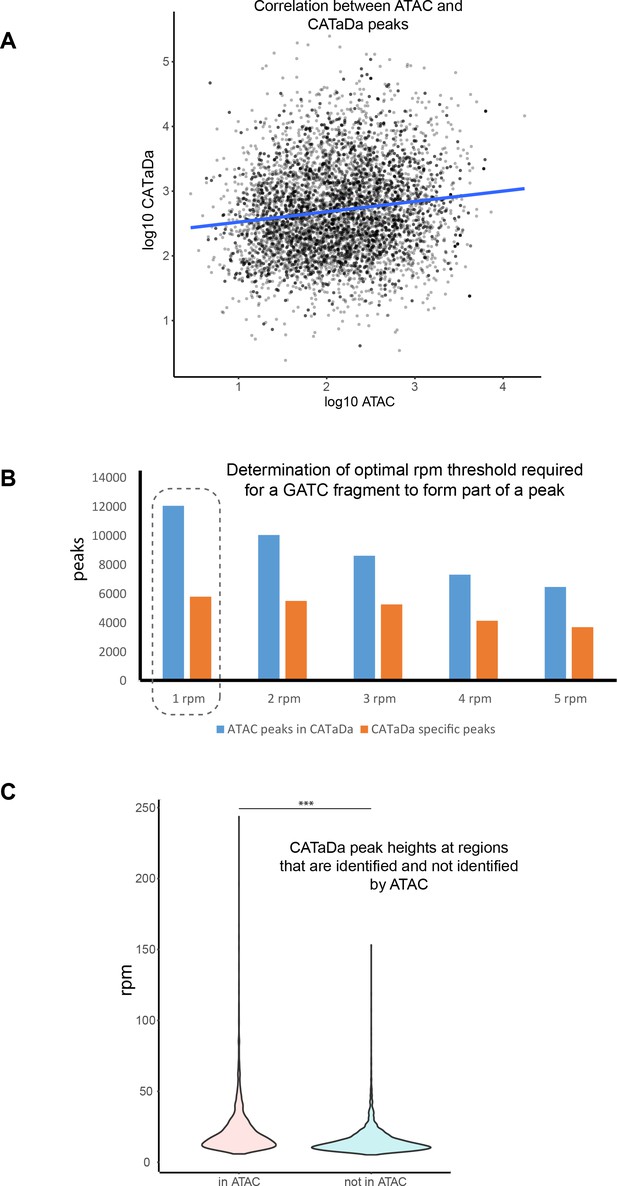
Further comparison of CATaDa and ATAC data.
(A) Correlation between peaks called in ATAC-seq and CATaDa. p<2.2 × 10−16, r2 = 0.137967. (B) Thresholding of peak calling at lower rpm values results in a higher proportion of ATAC peaks being identified whilst having little effect on the number of peaks identified, which are seen in CATaDa data, but not ATAC. (C) Peaks identified only in CATaDa are significantly smaller than those seen to overlap with ATAC peaks. p<1 × 10−16.
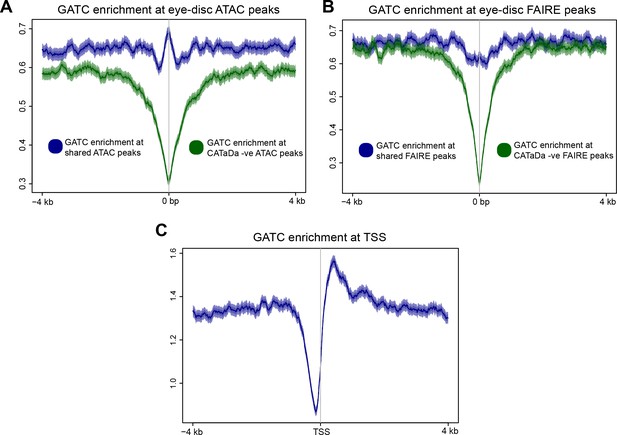
Frequency of GATC sites at various genomic features.
(A) Frequency of GATC sites at peaks identified by ATAC-seq in eye-discs. There is a clear reduction of GATC frequency at loci corresponding to peaks which were identified with ATAC, but not CATaDa (green), compared to peaks identified by both methods (blue). (B) Frequency of GATC sites at peaks identified by FAIRE-seq in eye-discs. There is a clear reduction of GATC frequency at loci corresponding to peaks which were identified with FAIRE, but not CATaDa (green), compared to peaks identified by both methods (blue). (C) Frequency of GATC sites around transcriptional start sites (TSS). Promoter regions have a depletion of GATC sequences.
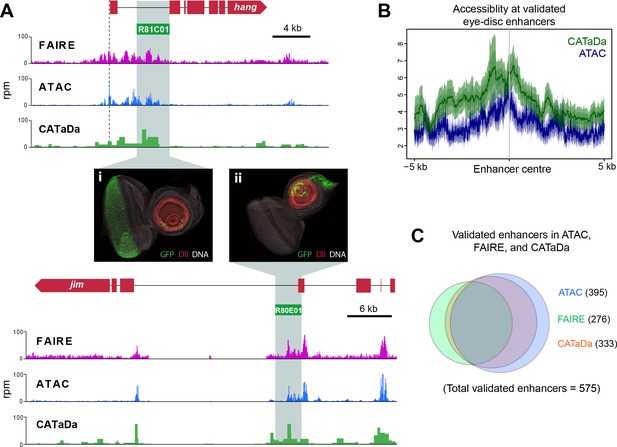
Identification of validated imaginal disc enhancers with CATaDa.
(A) Example loci showing data obtained by FAIRE, ATAC, and CATaDa. Peaks are broadly reproducible across techniques. Flylight enhancers with validated expression in eye imaginal discs coincide with peaks in all three datasets. Corresponding expression pattern is shown in (i) and (ii) (eye disc images obtained from the FlyLight database [http://flweb.janelia.org/cgi-bin/flew.cgi]). (B) Aggregation plot showing average signal of ATAC (blue) and Dam (green) at 575 FlyLight enhancers with validated eye imaginal disc expression. Both techniques show increased open chromatin at these regions. (C) Venn diagram of FlyLight enhancers identified in Dam accessibility profiling, ATAC, or FAIRE-seq. The majority of enhancers identified by either ATAC or FAIRE are also found in the Dam data. Dam enhancers overlap most with ATAC (305 shared between ATAC and Dam of 575 total FlyLight enhancers).
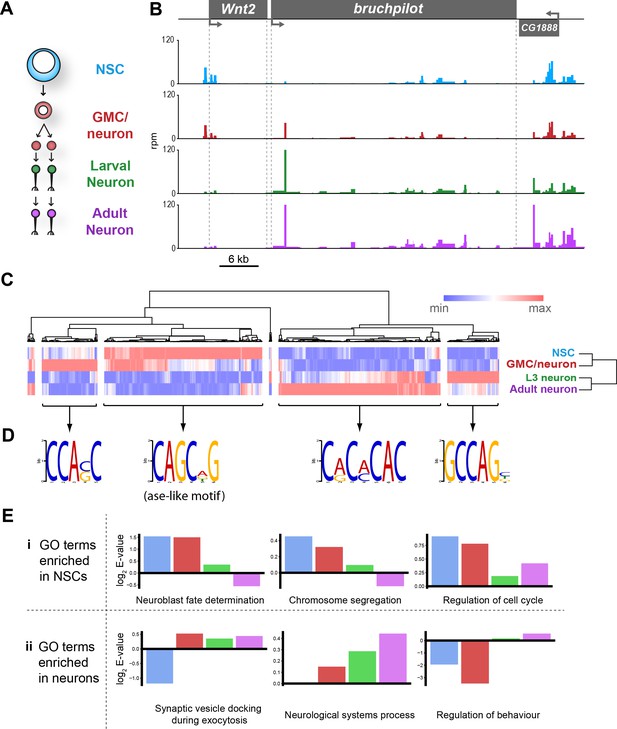
Chromatin accessibility of cell types in the CNS.
(A) Schematic of CNS lineage progression indicating cell types examined in this study. (B) Example profiles resulting from Dam expression in the CNS. Genomic region encompassing Wnt2 and bruchpilot genes is shown. Multiple open chromatin regions are dynamic across development. Y-axes = reads per million (rpm). (C) Clustering of differentially accessible regions in CNS lineages indicates two major groupings in which chromatin is most accessible in either stem cells or mature neurons. (D) Motif analysis using these sequences results in identification of expected motifs (e.g. ase E-box motif in stem cell accessible loci), as well as novel motifs. Most highly enriched motifs for each cluster shown. All motifs E-values < 1 × 10−5. (E) log2 enrichment scores for selected GO terms in individual cell types. Clear trends can be seen as development progresses. (NSC, GMC, L3 neuron, adult neuron - from left to right). (i) GO terms are either enriched in stem cells becoming less significant as the lineage progresses or (ii) vice versa.
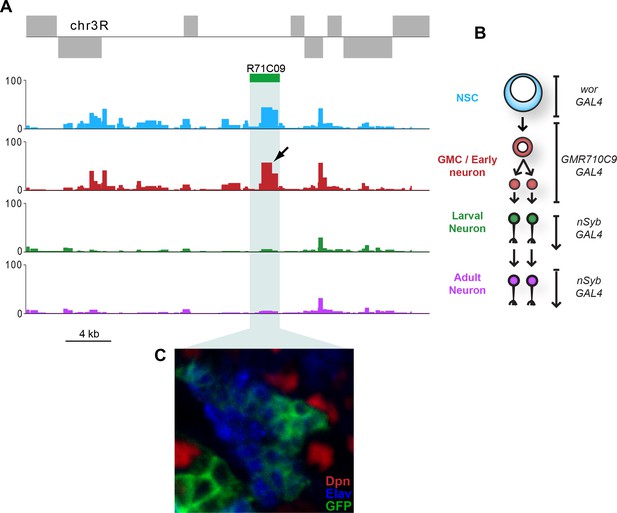
Chromatin accessibility in neural cell types demonstrating dynamic accessibility of R71C09 enhancer region - used to define GMC/immature neuron populations in this study.
(A) A clear peak can be identified within the R71C09 sequence which shows greatest accessibility in the GMCs and youngest populations of neurons in the lineage (arrow). (B) Schematic indicating cell types assayed in this study with GAL4 diver lines used to drive Dam expression. (C) Expression pattern of R71C09-GAL4/UAS-GFP in the larval CNS. GFP is most strongly detected in cells adjacent to the neuroblast (Dpn - Red). These cells include presumptive GMCs (Dpn negative, Elav negative), and immature neurons (Elav - Blue).
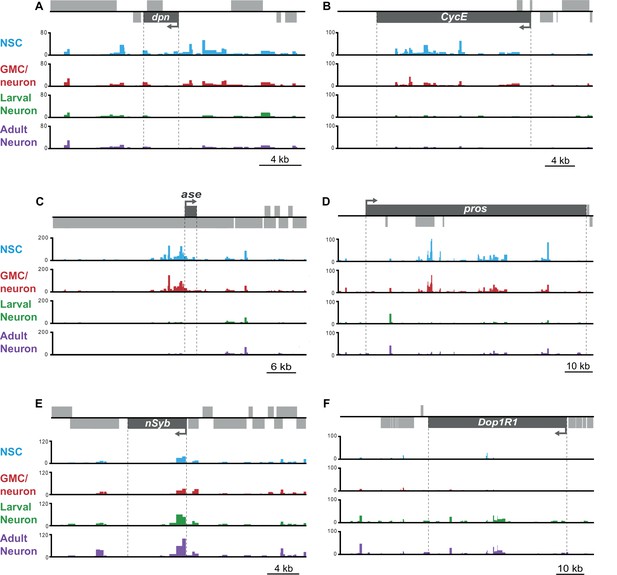
Example loci showing dynamic chromatin accessibility in neuronal cell-types.
Regions of open chromatin are enriched in progenitor cell types for (A) deadpan (dpn) (B) Cyclin E (CycE) (C) asense (ase), and (D) prospero (pros); whilst peaks of greater accessibility are apparent in differentiated neurons at the (E) neuronal Synaptobrevin (nSyb), and (F) Dopamine 1-like receptor 1 (Dop1R1) loci.
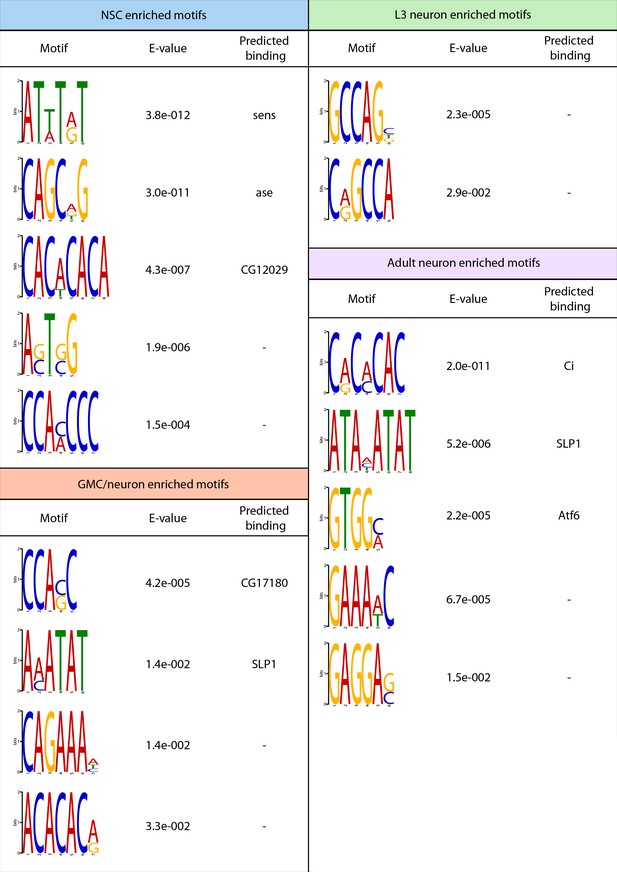
Top enriched motifs identified in regions of enhanced chromatin accessibility in neuronal cell types.
Top five enrched motifs are shown for each group, unless fewer than five total enriched motifs were identified. Predicted transcription factors binding to observed motifs are shown if detected.
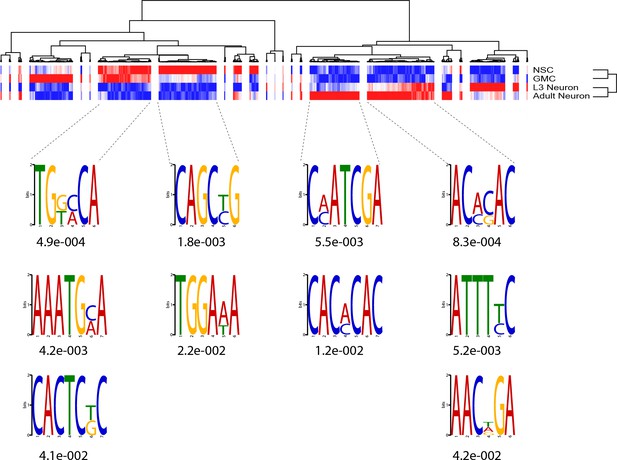
Further motifs enriched in cell types of the nervous system.
Further examination of sub-clusters reveals novel motifs enriched in different cell types of neurogenesis. The ase-like motif (CAGCNG) described in Figure 4D is enriched specifically in the NSC-specific cluster but not for regions that are accessible in both NSC and GMCs/neurons.
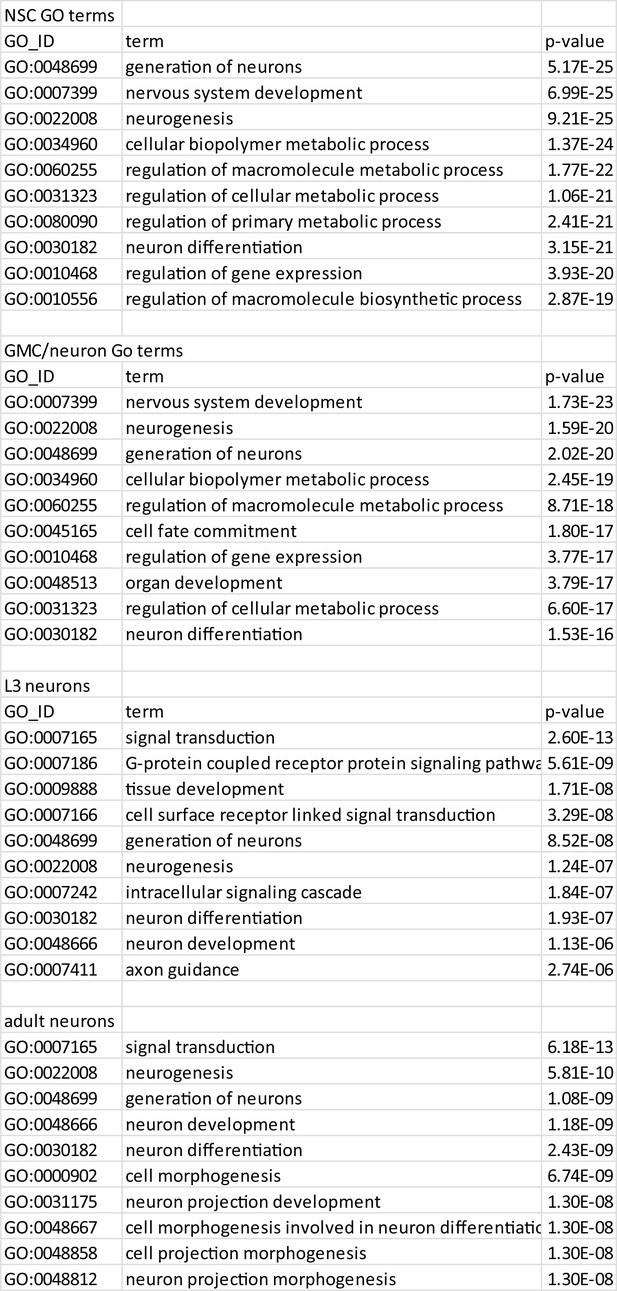
Top enriched GO terms for cell types of the CNS.
https://doi.org/10.7554/eLife.32341.014-
Figure 4—figure supplement 5—source data 1
Top enriched GO terms for cell types of the CNS as Excel spreadsheet.
- https://doi.org/10.7554/eLife.32341.015
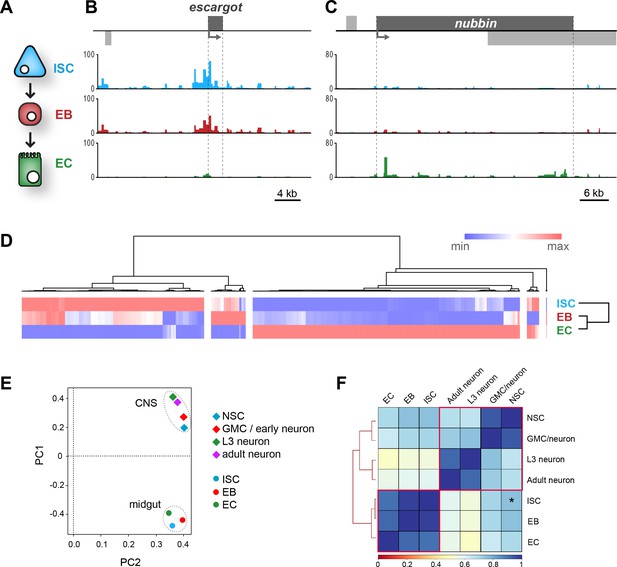
Dam chromatin accessibility profiling of cells in the adult midgut.
(A) Schematic of midgut lineage progression indicating cell types examined in this study. (B) Chromatin accessibility displays expected trends at the escargot locus, known to be expressed exclusively in ISCs and EBs, but not ECs. Upstream promoter region shows greatest chromatin accessibility in ISCs, compared to other cell types. Similarly, dynamic peaks are observed in both 3’ and 5’ distal regions (putative enhancer regions), which are absent in ECs. y-axes = reads per million (rpm). (C) Chromatin accessibility at the nubbin locus, known to be expressed exclusively in ECs. y-axes = reads per million (rpm). (D) Hierarchical clustering of differentially accessible regions in gut cell types. Major clusters are observed in which accessible chromatin is enriched specifically in either ISCs or ECs, whilst smaller clusters indicate fewer regions with up or down-regulated accessibility in EBs. (E) Principal component analysis (mean of all replicates) indicates distinct groupings of both lineages. (F) Correlation matrix (Spearman’s rank) of means of all cells in CNS and midgut lineages. Individual lineages denoted with red outline. Note relatively high correlation between NSC and ISC (Asterisk – R2 = 0.76), whilst NSC correlation with EC and adult neurons are comparable.
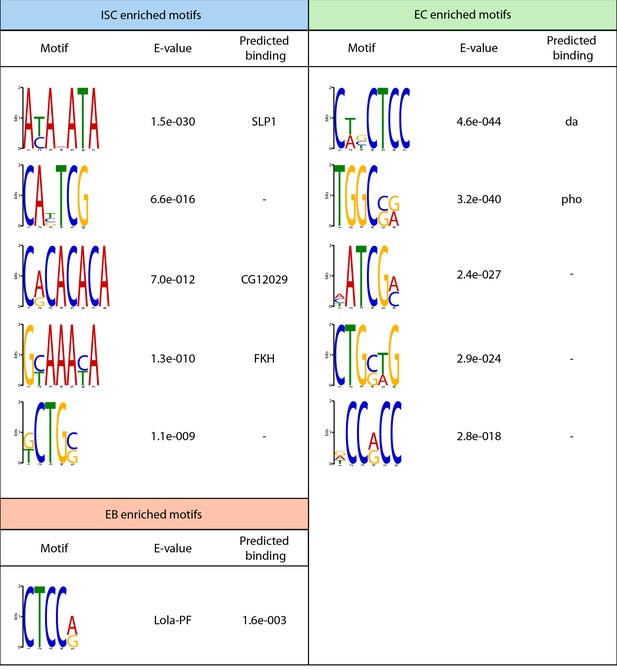
Top enriched motifs identified in regions of enhanced chromatin accessibility in midgut cell types.
Top five enrched motifs are shown for each group, unless fewer than five total enriched motifs were identified. Predicted transcription factors binding to observed motifs are shown if detected.
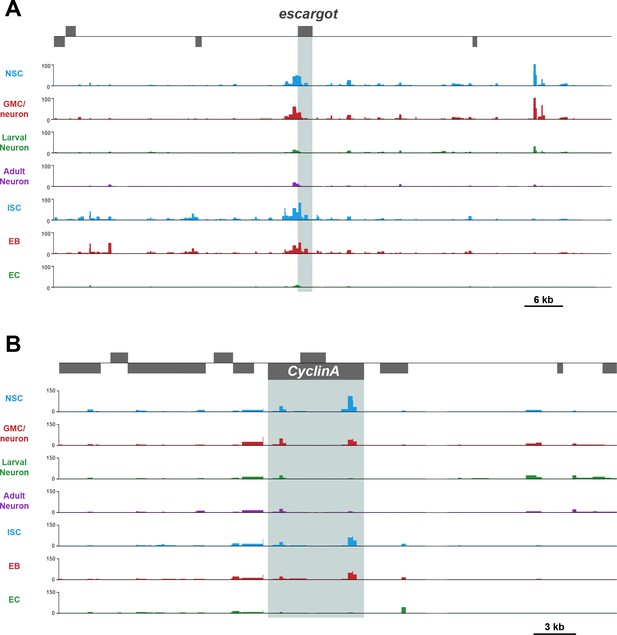
Example loci of growth related loci with similar chromatin accessibility in CNS and midgut development.
(A) escargot locus – chromatin is highly accessible in the ICS but reduced in the differentiated ECs, this is reflected in the CNS in which NSCs have accessible chromatin but not neurons. (B) Cyclin A locus shows a similar pattern to esgcargot.
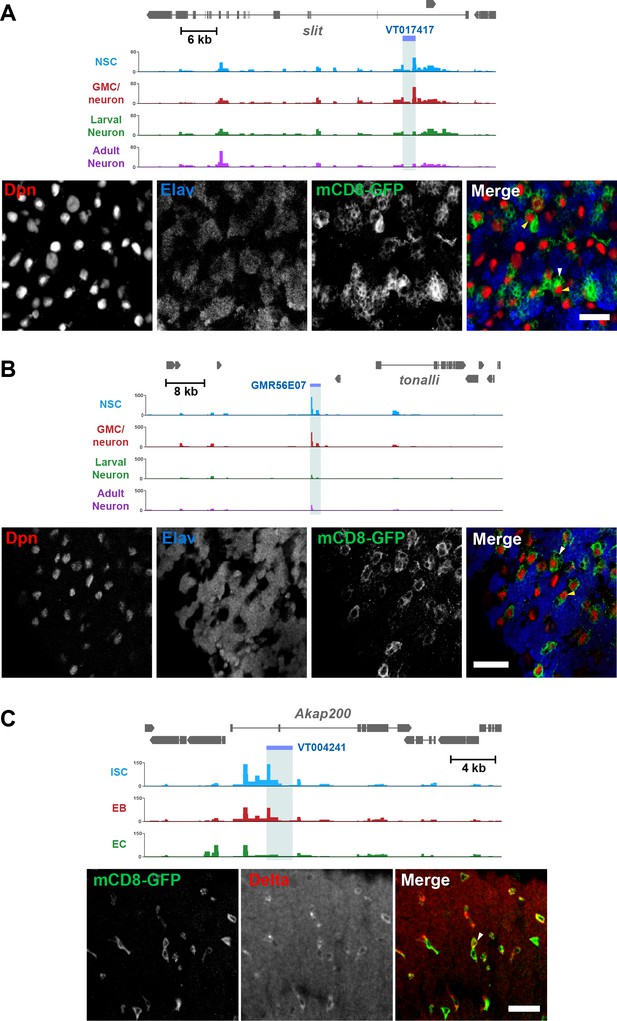
Identification of cell-type-specific enhancers from Dam accessibility data.
(A) Intronic putative enhancer region VT017417 within slit locus reveals expression of GFP reporter gene predominantly in GMCs (white arrow) and newly born neurons, as well as some NSCs (Dpn positive, yellow arrow). (B) Intergenic putative enhancer region GMR56E07 shows expression of GFP reporter gene predominantly in NSCs (yellow arrow), with some GMC expression (white arrow). (C) Intronic putative enhancer region VT004241 shows expression of GFP reporter predominantly in the ISCs (marked with Delta, white arrow). All scale bars = 20 µm.
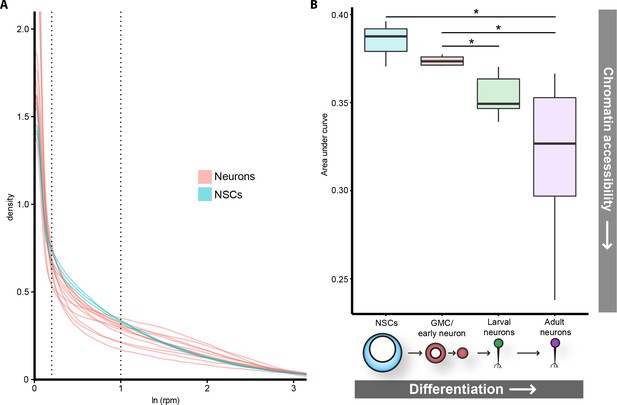
Global chromatin accessibility is reduced in differentiated neurons.
(A) Log transformed distribution of read counts at GATC fragments for neuronal (pink) or NSC replicates (blue). In addition to adult neuron data described in previous figures, CATaDa data for cholinergic, glutamatergic, and GABAergic adult neurons are included. NSC data include extra replicate from (Marshall et al., 2016). (B) Areas under curve for region bound by dotted lines in (A). Corresponding to ~1–3 rpm. Data include extra neuronal and NSC replicates shown in (A), as well as corresponding replicates at L3 for gutamatergic, GABAergic, and cholinergic neurons. Note that area under curve corresponds to proportion of GATC fragments having mapped reads within indicated range (i.e. NSCs have ~38% (median) of all GATC fragments within 1–3 rpm mapped reads, compared to ~32% for adult neurons. Results were considered significant at *p<0.05.
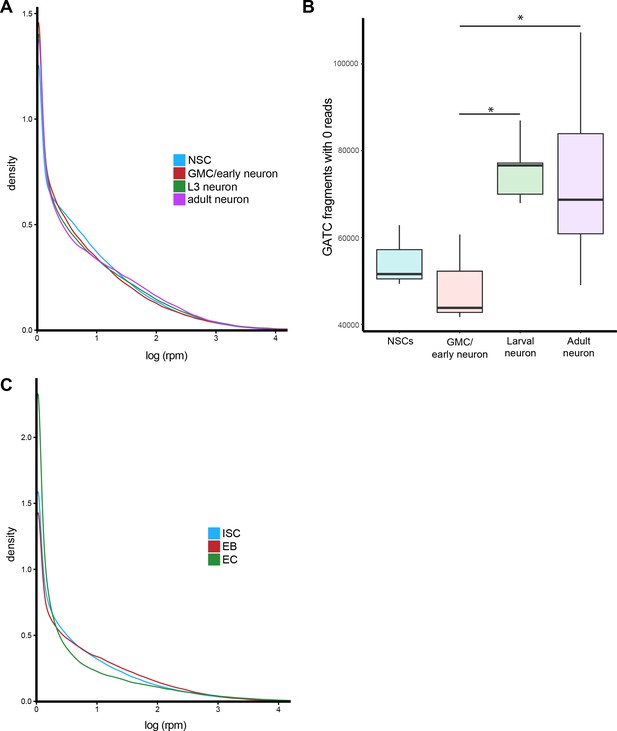
Average distribution of sequencing reads for CNS and midgut cell types.
(A) Mean distribution of reads for all cell types in the CNS. (B) Number of regions with zero mapped reads for cell types of the CNS. Results were considered significant at *p<0.05. (C) Mean distribution of reads for all cell types in the midgut.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32341.022





