Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability
Figures
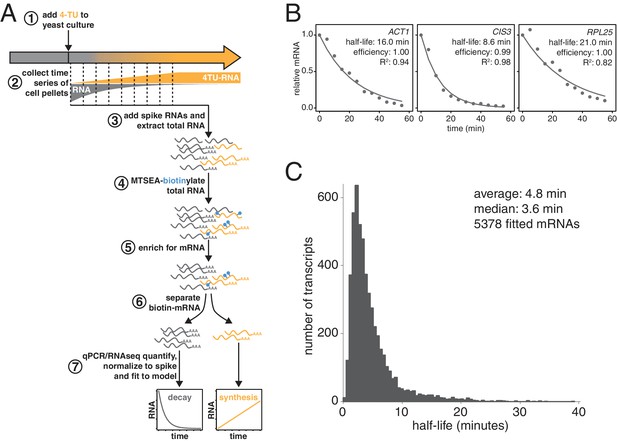
Improved mRNA stability measurements with metabolic labeling.
(A) Experimental scheme to measure mRNA stability and production by metabolic labeling. (B) Wild-type cells (KWY165) were subjected to the experiment described in (A) and decay rates for ACT1, CIS3 and RPL25 transcripts were determined by RT-qPCR. (C) mRNA samples in (B) were quantified by RNA-seq to determine mRNA stabilities across the transcriptome. Half-lives are plotted by frequency and each bin is 0.5 min wide.
-
Figure 1—source data 1
Source data for Figure 1B: decay kinetics of ACT1, CIS3 and RPL25 mRNAs.
- https://doi.org/10.7554/eLife.32536.005
-
Figure 1—source data 2
Source data for Figure 1C and Figure 1—figure supplement 1C: transcriptome wide decay data for two biological replicates of wild-type yeast cells grown in exponential phase.
- https://doi.org/10.7554/eLife.32536.006
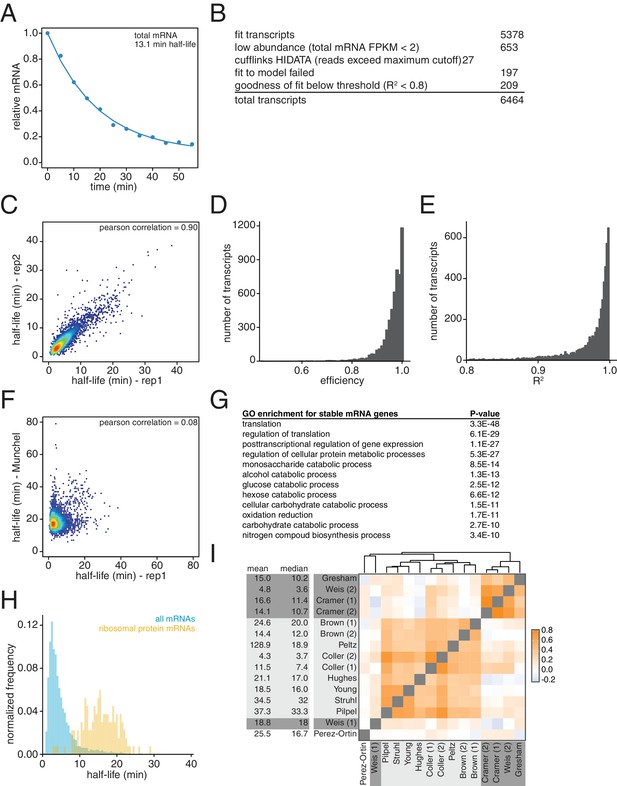
Characterization of transcriptome-wide mRNA stability profiling.
(A) Fragments per kilobase per million reads were summed for all mRNAs derived from the global stability profiling, normalized to the spike control and the time = 0 value, plotted and fit for half-life determination. (B) Numbers of transcripts that were successfully fit to the model and numbers of transcripts that failed to be measured for listed reasons. (C) Two biological replicates of the global mRNA stability profiling of wild-type cells (KWY165) were collected and transcript half-lives are plotted against each other. Red portions of the plot represent areas with dense datapoints and blue portions of the plot represent areas with sparse datapoints. (D) Distribution of efficiency parameters for the experiment described in Figure 1C. Each bin is 0.01 units wide. (E) Distribution of R2 values for the experiment described in Figure 1C. Each bin is 0.002 units wide. (F) Scatter plot of half-life values measured in this study compared to the values reported in Munchel et al. (2011). Colors represent density of datapoints as in Figure 1—figure supplement 1C. (G) Functional enrichment of long-lived transcripts as defined as transcripts having a half-life longer than one standard deviation greater than the mean half-life. (H) Half-life distributions of all 137 ribosomal protein-encoding mRNA half-lives (yellow) compared to the entire transcriptome (blue) normalized to the size of each transcript group. Each bin is 0.5 min wide. (I) Spearman correlation coefficients were computed for pairs of mRNA stability datasets and plotted. Orange represents positive correlation and blue represents negative correlation. Text background colors represents experimental methodology: metabolic labeling in dark gray, transcriptional shutoff in light gray and other in white. The datasets were hierarchically clustered based on Euclidian distances. The datasets were derived from the following: Gresham (Neymotin et al., 2014), Weis (2) [this study], Cramer (Shyu et al., 1991; Miller et al., 2011), Cramer (Muhlrad and Parker, 1992; Sun et al., 2012), Brown (1) and (Muhlrad and Parker, 1992; Wang et al., 2002), Peltz (Duttagupta et al., 2005), Coller (1) and (Muhlrad and Parker, 1992; Presnyak et al., 2015), Hughes (Grigull et al., 2004), Young (Holstege et al., 1998), Struhl (Geisberg et al., 2014), Pilpel (Shalem et al., 2008), Weis (Shyu et al., 1991; Munchel et al., 2011), Perez-Ortin (Pelechano and Pérez-Ortín, 2010).
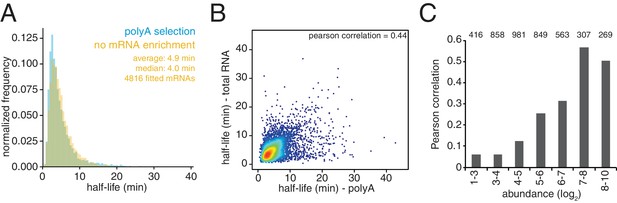
Comparison of mRNA stabilities in the presence or absence of mRNA enrichment.
(A) The experiment described in Figure 1C was performed in the absence of oligo-dT selection (no mRNA enrichment). Half-lives are plotted by frequency against the data derived from the experiment in Figure 1C (polyA selection) and the width of each bin is 0.5 min. (B) Scatter plot of half-life values from (A) and Figure 1C. Colors represent density of datapoints as in Figure 1—figure supplement 1C. (C) Data were binned by abundance and the pearson correlation for each of these subgroups between the data from A and Figure 1C is plotted. The population of each bin is indicated by the number above each bar.
-
Figure 1—figure supplement 2—source data 1
Source data for Figure 1—figure supplement 2A–B: transcriptome wide decay data prepared without polyA selection.
- https://doi.org/10.7554/eLife.32536.007
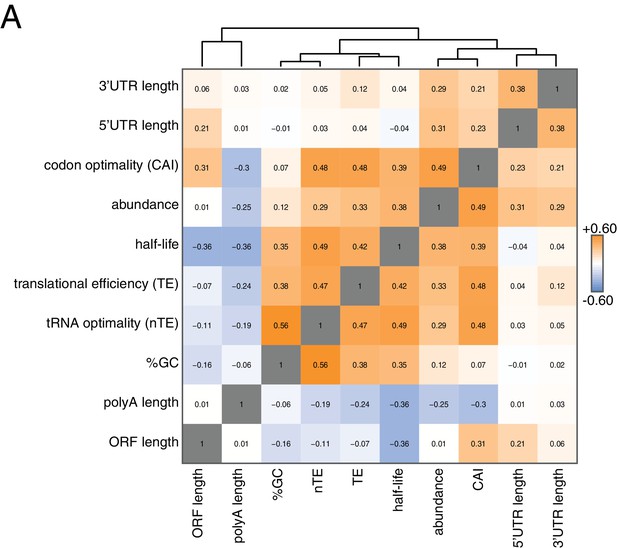
Correlation of mRNA features.
(A) Spearman rank correlation coefficients were computed for pairs of mRNA parameters of stability (half-life), translation efficiency (TE), polyA tail length, codon optimality (CAI), tRNA optimality (nTE), abundance, UTR lengths, GC content and ORF length and plotted as a heatmap. Datasets were hierarchically clustered based on Euclidian distances. Orange represents positive correlation and blue represents negative correlation. Correlations between identical datasets are colored in gray. See Supplementary file 1 for sources of genome wide data.
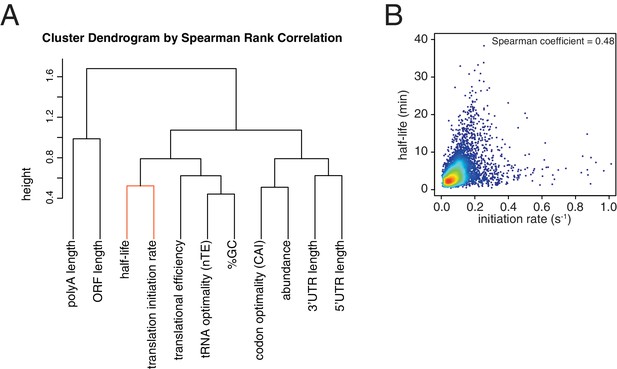
Correlation between translation initiation rate and aspects of mRNA structure and function.
(A) Dendrogram of the Spearman rank correlation coefficients between transcriptome-wide datasets. Clustering was based on Euclidian distances. The close clustering between half-life and translation initiation rate is highlighted in orange. (B) Scatter plot of half-life values against estimated translation initiation rates. Color indicates density of datapoints as in Figure 1—figure supplement 1C.
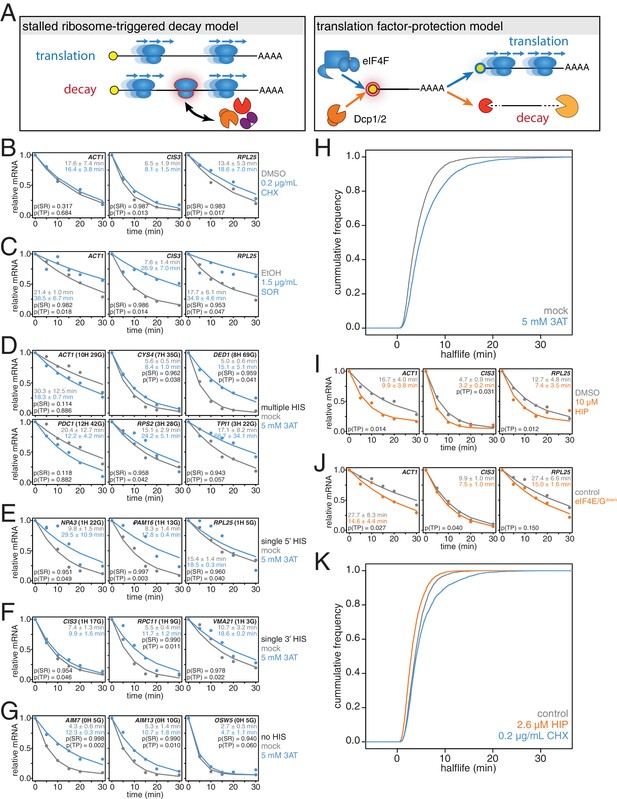
mRNAs are stabilized by slowly elongating ribosomes and destabilized when translation initiation is inhibited.
(A) Cartoon depictions of the stalled ribosome-triggered decay and translation factor-protection models. (B) Wild-type cells (KWY165) were subjected to mRNA stability profiling immediately after addition of 0.1% DMSO or 0.2 μg/mL cycloheximide in 0.1% DMSO. Data on ACT1, CIS3 and RPL25 mRNAs were collected and plotted. See Figure 3—figure supplement 4A for biological replicates. P-values are computed using a one-sided paired t-test for both the stalled ribosome-triggered decay model (p(SR)) as well as the translation factor-protection model (p(TP)). P-values less than 0.05 are significant. (C) Wild-type cells (KWY165) were subjected to mRNA stability profiling 33 min after addition of 0.1% ethanol or 1.5 μg/mL sordarin in 0.1% ethanol (note that this is the timepoint when a growth defect is manifested, see Figure 3—figure supplement 1C). Data were collected, analyzed and plotted as in Figure 3B. See Figure 3—figure supplement 4B for biological replicates. (D–G) HIS3 gcn2∆ cells (KWY7337) were subjected to mRNA stability profiling immediately after non-addition (mock) or addition of 5 mM 3AT. Data were collected, analyzed and plotted as in Figure 3B. See Figure 3—figure supplement 4C for biological replicates. (H) mRNA samples collected from the experiment described in Figure 3D–G were subjected to global mRNA stability profiling. Cumulative frequencies of transcript half-life are plotted. (I) Wild-type cells (KWY165) were subjected to mRNA stability profiling immediately after addition of 0.1% DMSO or 10 μM hippuristanol. Data were collected, analyzed and plotted as in Figure 3B. p-values were not computed for the stalled ribosome-triggered decay model as this model does not make a clear prediction as to how mRNA stability is affected when translation initiation is perturbed. See Figure 3—figure supplement 5A for biological replicates. (J) pGPD1-LexA-EBD-B112 CDC33-3V5-IAA7 pRS425 cells (KWY7336: control) and pGPD1-LexA-EBD-B112 CDC33-3V5-IAA7 pGPD1-OsTIR1 pRS425-p4xLexOcyc1-CDC33∆CAP cells (KWY7334: eIF4E/Gdown) were grown in CSM-LEU-0.5xURA pH5.5 media and subjected to mRNA stability profiling immediately after addition of 10 nM β-estradiol, 100 μM 3-indoleacetic acid and 4 μM IP6. Data were collected, analyzed and plotted as in Figure 3I. See Figure 3—figure supplement 5B for biological replicates. (K) Wild-type cells (KWY165) were subjected to global mRNA stability profiling immediately after addition of 0.1% DMSO (gray) or 2.6 μM hippuristanol (orange) or 0.2 μg/mL cycloheximide (blue). Cumulative frequencies of transcript half-life are plotted.
-
Figure 3—source data 1
Source data for Figure 3B, C, D–G, I and J: decay kinetics of selected mRNAs in cells treated with translational inhibitors.
- https://doi.org/10.7554/eLife.32536.014
-
Figure 3—source data 2
Source data for Figure 3H: Transcriptome wide decay data for cells treated or mock treated with 3AT.
- https://doi.org/10.7554/eLife.32536.015
-
Figure 3—source data 3
Source data for Figure 3K: Transcriptome wide decay data for cells treated with DMSO, cycloheximide or hippuristanol.
- https://doi.org/10.7554/eLife.32536.016
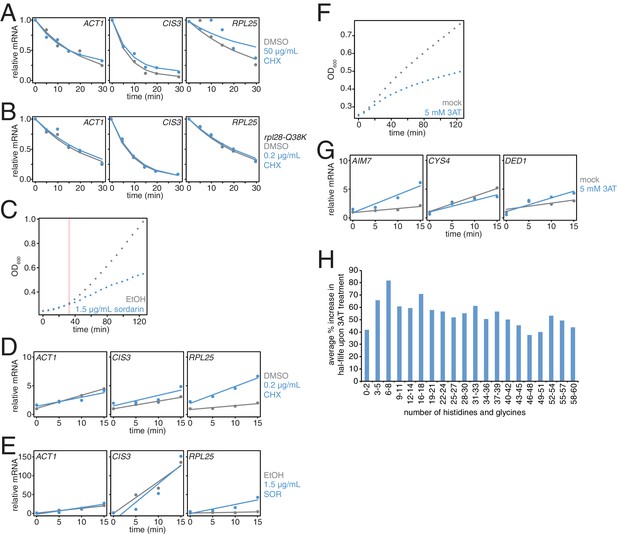
Effects of translation elongation inhibitors on mRNA stability and cell growth.
(A) Wild-type cells (KWY165) were subjected to mRNA stability profiling immediately after addition of 0.1% DMSO or 50 μg/mL cycloheximide in 0.1% DMSO. Data were collected and plotted as in Figure 3B. (B) rpl28-Q38K cells (KWY7335) were subjected to mRNA stability profiling immediately after addition of 0.1% DMSO or 0.2 μg/mL cycloheximide in 0.1% DMSO. Data were collected and plotted as in Figure 3B. (C) Wild-type cells (KWY165) were treated with 0.1% ethanol (gray) or 1.5 μg/mL sordarin in 0.1% ethanol (blue) and growth by absorbance at 600 nm was monitored. The horizontal orange line marks t = 33 min where the growth rates diverge. (D) Thio-uracil containing mRNAs from the experiment described in Figure 3B were measured and levels were plotted. RNA levels were normalized to a 4TU-labeled hSRP1 spike RNA and to the time = 0 value for the mock (DMSO) treated sample. (E) Thio-uracil containing mRNAs from the experiment described in Figure 3C were measured and levels were plotted. RNA levels were normalized to a 4TU-labeled hSRP1 spike RNA and to the time = 0 value for the mock (EtOH) treated sample. (F) Wild-type cells (KWY165) were treated with 5 mM 3AT (blue) or untreated (gray) and growth by absorbance at 600 nm was monitored. (G) Thio-uracil containing AIM7, CYS4 and DED1 mRNAs from the experiment described in Figure 3C–E were measured and levels were plotted. RNA levels were normalized to a 4TU-labeled hSRP1 spike RNA and to the time = 0 value for the mock (DMSO) treated sample. (H) Fold increase in mRNA half-life values when cells are treated with 5 mM 3AT (derived from the experiment described in Figure 3H) were computed for groups of mRNAs binned by their histidine and glycine codon contents.
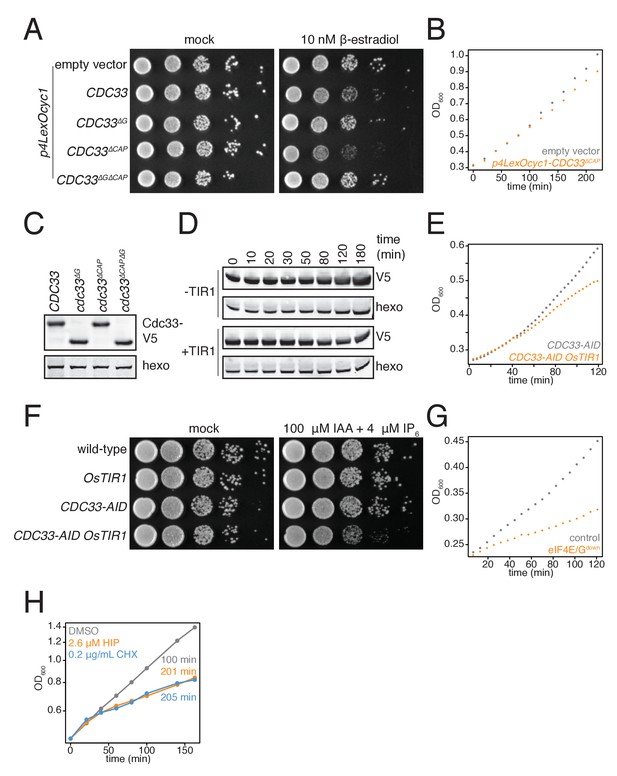
Biological replicates of experiments described in Figure 3B–G.
(A) pGPD1-LexA-B112 cells harboring the following vectors: pRS425 (KWY7327 empty vector), pRS425-p4LexOCYC1-CDC33 (KWY7329), pRS425-p4LexOCYC1-CDC33∆G (KWY7330), pRS425-p4LexOCYC1-CDC33∆CAP (KWY7331) and pRS425-p4LexOCYC1-CDC33∆CAP∆G (KWY7332) were spotted on CSM-LEU and CSM-LEU + 10 nM β-estradiol media and incubated at 30°C for 36 hr. The first spot represents growth of approximately 6 × 104 cells and each subsequent spot is a 10 fold serial dilution. (B) pGDP1-LexA-B112 cells harboring pRS425 (KWY7327 empty vector, gray) or pRS425-p4LexOCYC1-CDC33∆CAP (KWY7331, orange) were grown to mid exponential phase, treated with 10 nM β-estradiol and growth was monitored by absorbance at 600 nm. (C) pGDP1-LexA-B112 cells harboring the following vectors: pRS425-p4LexOCYC1-CDC33-3V5 (KWY7328), pRS425-p4LexOCYC1-CDC33∆G-3V5 (KWY7338), pRS425-p4LexOCYC1-CDC33∆CAP-3V5 (KWY7339) and pRS425-p4LexOCYC1-CDC33∆CAP∆G-3V5 (KWY7340) were grown to mid exponential phase in CSM-LEU media, treated with 10 nM β-estradiol for 20 min and then harvested for western blot analysis. Hexokinase was used as a loading control. (D) CDC33-IAA7-3V5 cells expressing OsTIR1 (KWY7326) or not expressing OsTIR1 (KWY7325) were grown to mid exponential phase in CSM pH5.5 media, treated with 100 μM 3-indoleacetic acid and 4 μM IP6 and then harvested at the indicated timepoints for western blot analysis. Hexokinase was used as a loading control. (E) CDC33-IAA7-3V5 cells expressing OsTIR1 (KWY7326, orange) or not expressing OsTIR1 (KWY7325, gray) were grown to mid exponential phase in CSM pH5.5 media, treated with 100 μM 3-indoleacetic acid and 4 μM IP6 and growth by absorbance at 600 nm was monitored. (F) Wild-type (KWY165), pGPD1-OsTIR1 (KWY7333), CDC33-IAA7-3V5 (KWY7325) and pGPD1-OsTIR1 CDC33-IAA7-3V5 (KWY7326) cells were spotted on YePD pH5.5 and YePD pH5.5+100 μM 3-indoleacetic acid and 4 μM IP6 media and incubated at 30°C for 36 hr. The first spot represents growth of approximately 6 × 104 cells and each subsequent spot is a 10 fold serial dilution. (G) pGPD1-LexA-B112 CDC33-IAA7-3V5 pRS425 (KWY7336 ‘control’, gray) and pGPD1-LexA-B112 pGPD1-OsTIR1 CDC33-IAA7-3V5 pRS425-p4LexOCYC1-CDC33∆CAP (KWY7334 ‘eIF4E/Gdown’, orange) cells were grown in CSM –LEU pH5.5 to mid exponential phase, treated with 10 nM β-estradiol, 100 μM 3-indoleacetic acid and 4 μM IP6 and growth by absorbance at 600 nm was monitored. (H) Wild-type cells (KWY165) were treated with 0.1% DMSO (gray) or 2.6 μM hippuristanol (orange) or 0.2 μg/mL cycloheximide (blue) and growth was monitored by absorbance at 600 nm. The y-axis is in log scale. The values are the computed doubling times.
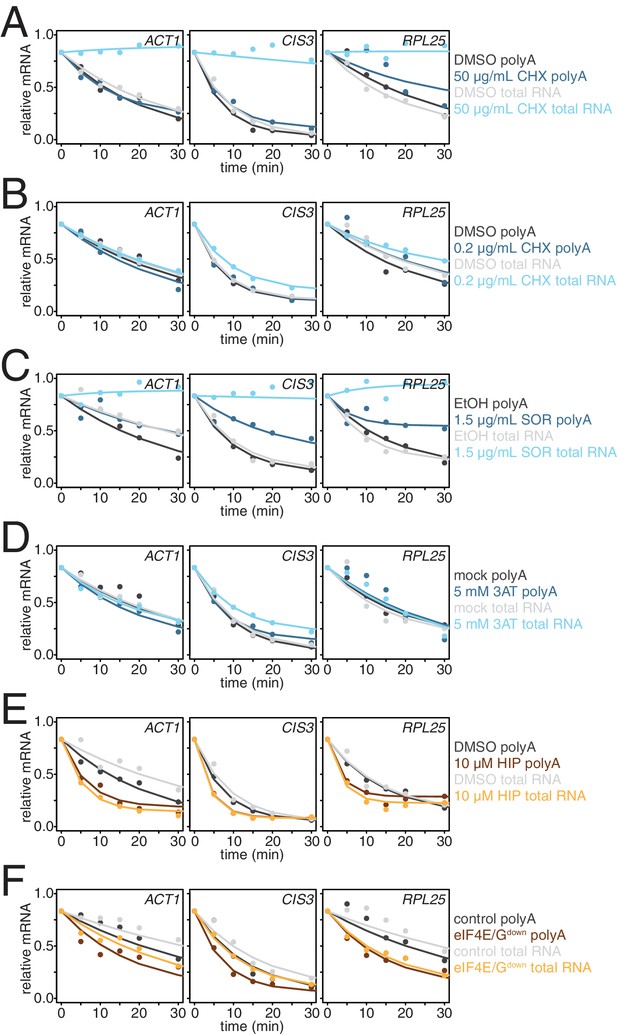
Effects of different modes of translation inhibition on mRNA stability in the absence of mRNA enrichment.
(A) Decay profiling of ACT1, CIS3 and RPL25 transcripts was performed for the experiment described in Figure 3—figure supplement 1A in the absence of oligo-dT bead selection (total RNA) and were fitted and plotted against measurements derived in the presence of oligo-dT selection (polyA). (B) Decay profiling as described in (A) was performed for the experiment described in Figure 3B. (C) Decay profiling as described in (A) was performed for the experiment described in Figure 3C. (D) Decay profiling as described in (A) was performed for the experiment described in Figure 3D–G. (E) Decay profiling as described in (A) was performed for the experiment described in Figure 3I. (F) Decay profiling as described in (A) was performed for the experiment described in Figure 3J.
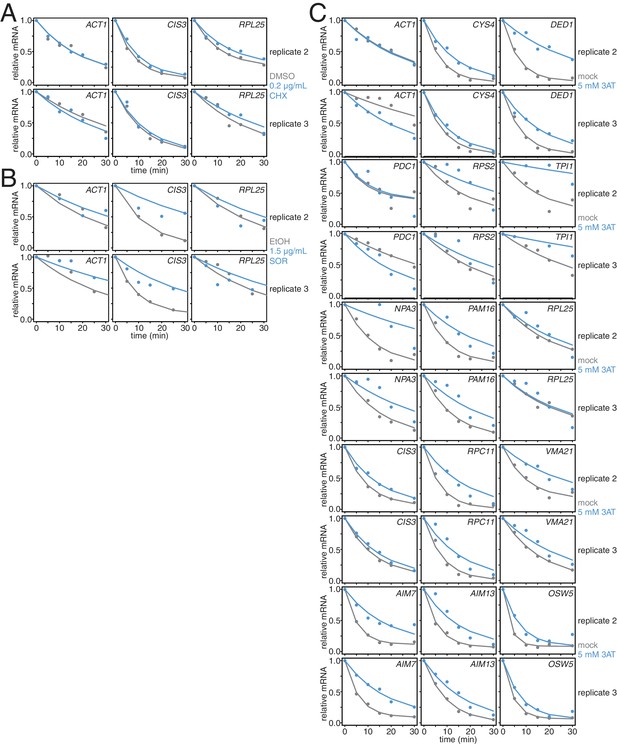
Biological replicates of experiments described in Figure 3I–J.
(A) Two additional biological replicates of the experiment described in Figure 3B. (B) Two additional biological replicates of the experiment described in Figure 3C. (C) Two additional biological replicates of the experiment described in Figure 3D–G.
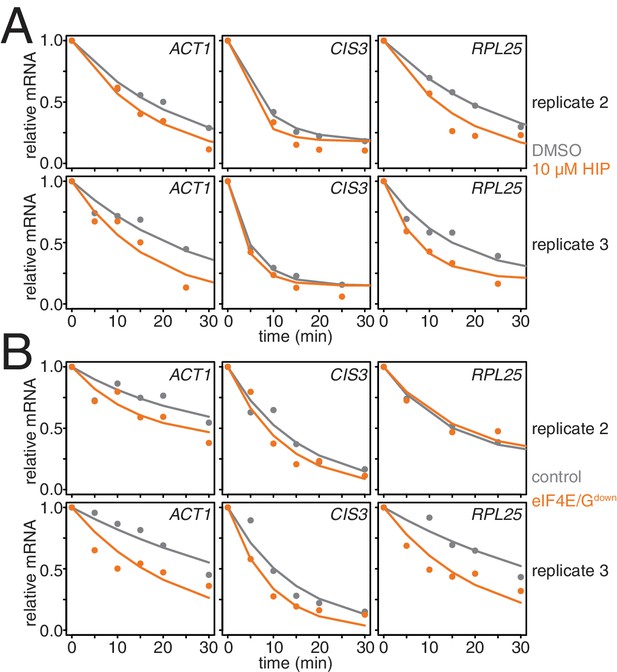
Characterization of conditional mutants of translation initiation.
(A) Two additional biological replicates of the experiment described in Figure 3I. (B) Two additional biological replicates of the experiment described in Figure 3J.
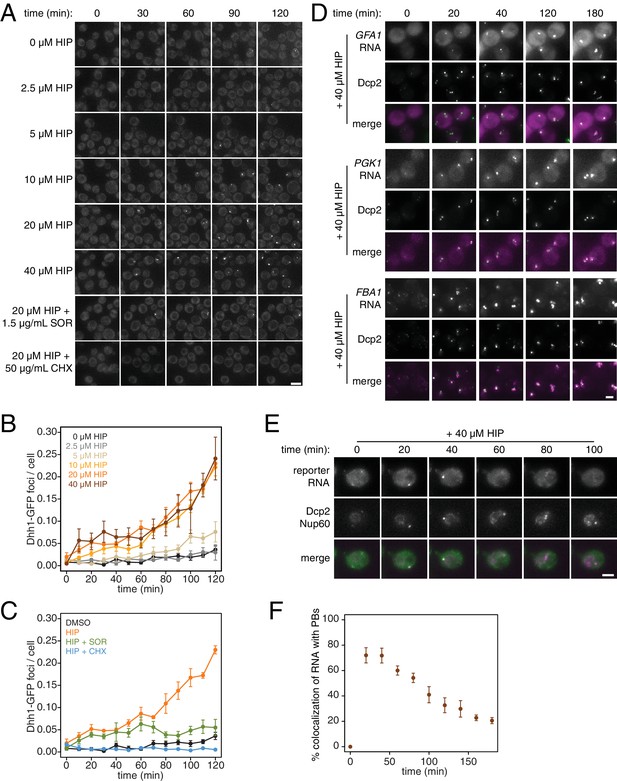
PB formation is stimulated by inhibiting translation initiation and blocked when translation elongation is inhibited.
(A) Dhh1-GFP, Dcp2-mCherry expressing cells (KWY5948) were grown to exponential phase and then treated with 0.1% DMSO, the indicated concentration of hippuristanol or co-treated with the indicated concentration of hippuristanol and either sordarin or cycloheximide. Images were acquired every 5 min using a wide-field microscope and the images were deconvolved. Shown are maximum projections of 8 z-stacks at a distance of 0.4 μm apart. Scale bar: 5 μm. (B–C) Number of Dhh1-GFP foci per cell from experiment in (A) was counted using Diatrack 2.5 particle tracking software. Error bars represent SEM (n = 3 biological replicates,>300 cells counted per experiment). (D) Dcp2-GFP, PP7CP-mKate2 expressing cells carrying PP7sl tagged copies of GFA1 (KWY7246), PGK1 (KWY6963) or FBA1 (KWY7245) were treated with 40 μM hippuristanol and immediately imaged. Images where acquired every 20 min using a wide-field microscope. Shown are maximum projections of 8 z-stacks at a distance of 0.5 μm apart. Scale bar: 2 μm. (E) Dcp2-mCherry, Nup60-3xmKate2, PP7CP-GFP expressing cells carrying a synthetic 3xGST-24xPP7sl under β-estradiol inducible control (KWY7227) were grown to mid-exponential phase, treated with 400 nM β-estradiol for 40 min and then transferred to media lacking β-estradiol and containing 40 μM hippuristanol and immediately imaged (see Figure 4—figure supplement 1D for the no hippuristanol control). Images were acquired every 20 min using a wild-field microscope. Shown are maximum projections of 8 z-stacks at a distance of 0.5 μm apart. Scale bar: 5 μm. For DMSO control images, see Figure 4—figure supplement 1D. (F) Images acquired in (E) were quantified for the colocalization of PP7CP-GFP foci with Dcp2-mCherry foci using FIJI software. Error bars represent SEM (n = 4 biological replicates,>120 PBs counted per timepoint).
-
Figure 4—source data 1
Source data for Figure 4B, C and F: accumulation kinetics of P-bodies and decay of RNA in P-bodies in cells treated with translational inhibitors.
- https://doi.org/10.7554/eLife.32536.021
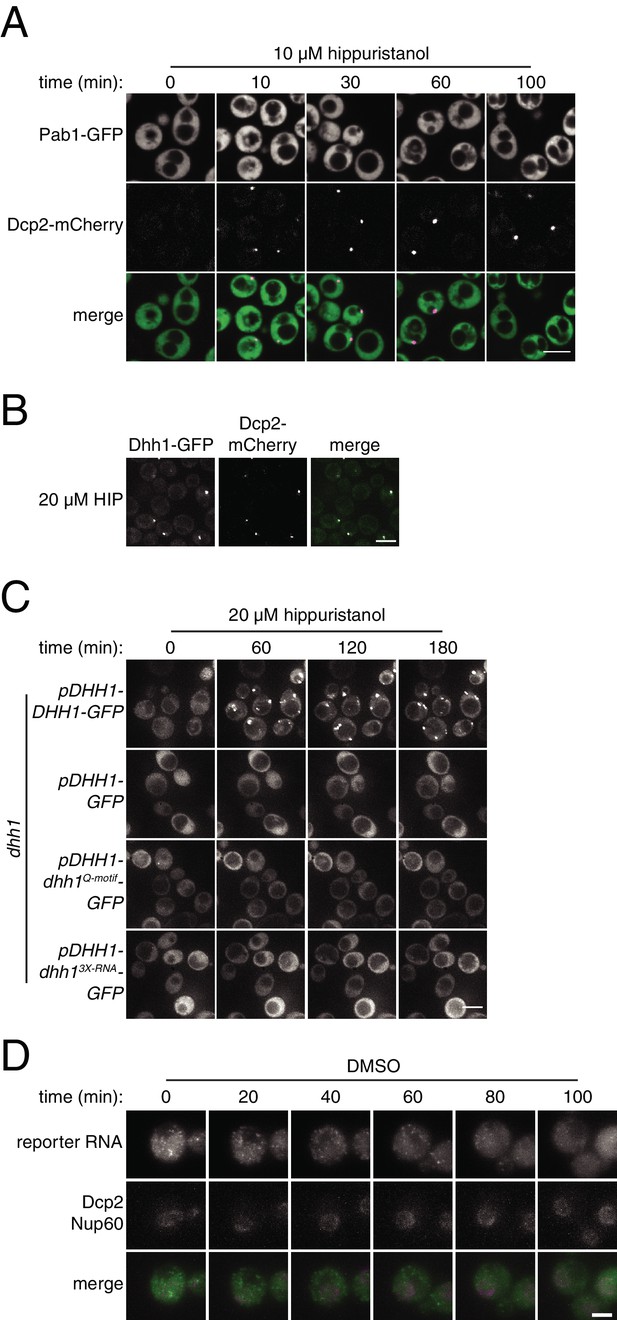
Characterization of P-body and stress-granule behavior in response to translation initiation inhibition.
(A) Pab1-GFP, Dcp2-mCherry expressing cells (KWY6554) were grown to exponential phase, treated with 10 µM hippuristanol and imaged using a confocal microscope (n = 2 biological replicates). Scale bar: 5 μm. (B) Dhh1-GFP, Dcp2-mCherry expressing cells (KWY5948) were grown to exponential phase and then treated with 20 μM hippuristanol for 120 min. Imaging and image processing was performed as in Figure 4A (n = 4 biological replicates) Scale bar: 5 μm. Note that this image is from the same experiment depicted in Figure 4A. (C) dhh1∆ cells expressing Dcp2-mCherry carrying plasmids pDHH1-DHH1-GFP (KWY3238), pDHH1-GFP (KWY5246), pDHH1-dhh1Q-motif-GFP (KWY5244) or pDHH1-dhh13X-RNA-GFP (KWY5242) were grown to exponential phase, treated with 20 μM hippuristanol and samples were imaged at the indicated times. Imaging and image processing was performed as in Figure 4A (n = 3 biological replicates). Scale bar: 5 μm. (D) Dcp2-mCherry, Nup60-3xmKate2, PP7CP-GFP expressing cells carrying a synthetic 3xGST-24xPP7sl under β-estradiol inducible control (KWY7227) were grown to mid-exponential phase, treated with 400 nM β-estradiol for 40 min and then transferred to media lacking β-estradiol and containing 0.4% DMSO and immediately imaged. Imaging and image processing was performed as in Figure 4E (n = 4 biological replicates). Scale bar: 2 μm.
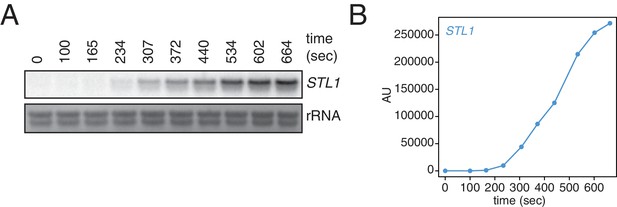
Kinetics of STL1 mRNA induction.
(A) Wild-type cells (KWY165) were grown to exponential phase, subjected to a 0.4 M NaCl salt shock and samples were collected at the indicated times for northern blot analysis. (B) Quantification of data in (A). STL1 mRNA levels were corrected to background and also to rRNA levels.
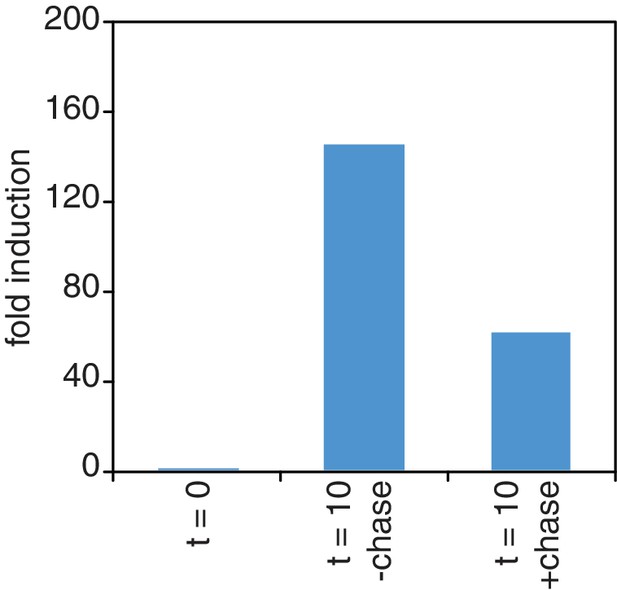
Chase inefficiency as revealed by STL1 induction.
Wild-type cells (KWY165) were grown to OD600 = 0.2, labeled with 0.2 mM 4TU for 2 hr and then washed out of the labeling media. Half of the cells were returned to the same media and the other half were resuspended in chase media containing 19.6 mM uracil. Both cultures were immediately subjected to 0.4 M NaCl salt shock and samples were collected 10 min after the salt shock. Thio-labeled mRNAs were purified and abundance of STL1 thio-mRNA was determined by qPCR, normalized to a thio-labeled spike (srp1α (Hs)) and then normalized to the levels of STL1 in pre-salt shocked cells (t = 0).
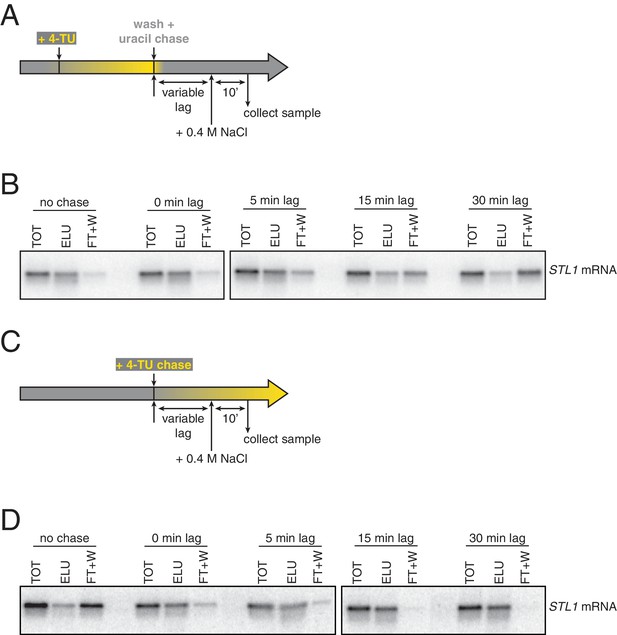
Analysis of chase efficiencies as a function of lag time and labeling scheme.
(A) 4TU-pulse/uracil-chase labeling scheme and experimental setup for (B). (B) Wild-type cells (KWY165) were grown to exponential phase, the experiment outlined in (A) was performed and levels of STL1 mRNA were determined by northern blot. (C) 4TU-chase labeling scheme and experimental setup for (D). (D) Wild-type cells (KWY165) were grown to exponential phase, the experiment outlined in (C) was performed and levels of STL1 mRNA were determined by northern blot.
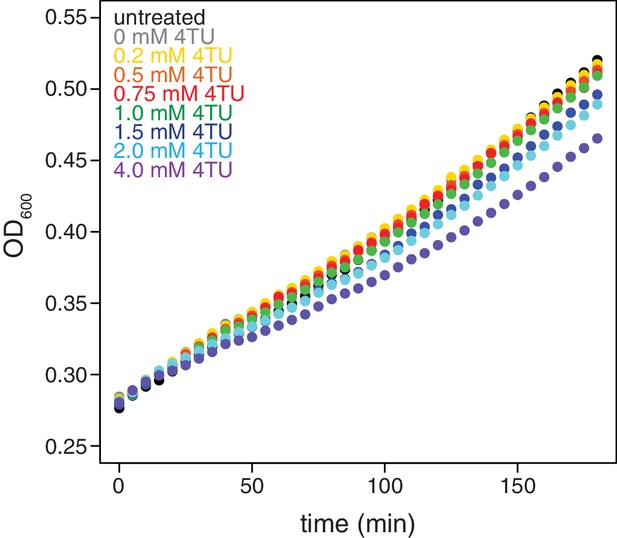
Effects of 4TU on cell growth.
Wild-type cells (KWY165) were grown to exponential phase, treated with the indicated concentration of 4TU and growth was monitored by absorbance at 600 nm.
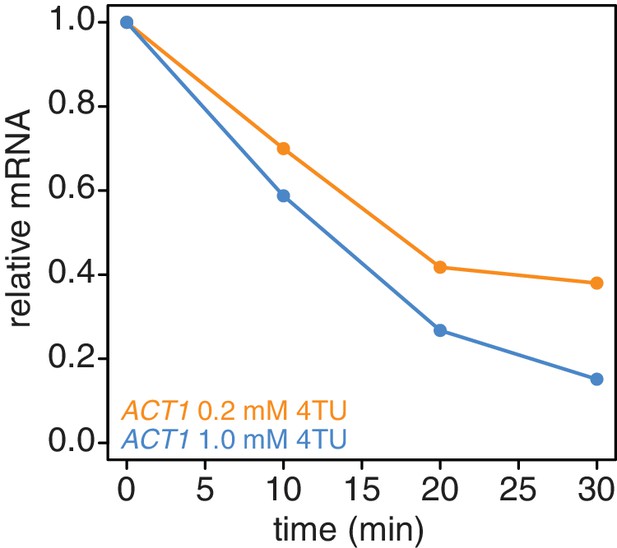
Increasing the 4TU concentration during the chase improves the subtraction of newly synthesized mRNAs.
Wild-type cells (KWY165) were subjected to the 4TU-chase protocol with either 0.2 mM 4TU in the chase (orange) or 1 mM 4TU (blue) and the unlabeled ACT1 mRNA levels were quantified.
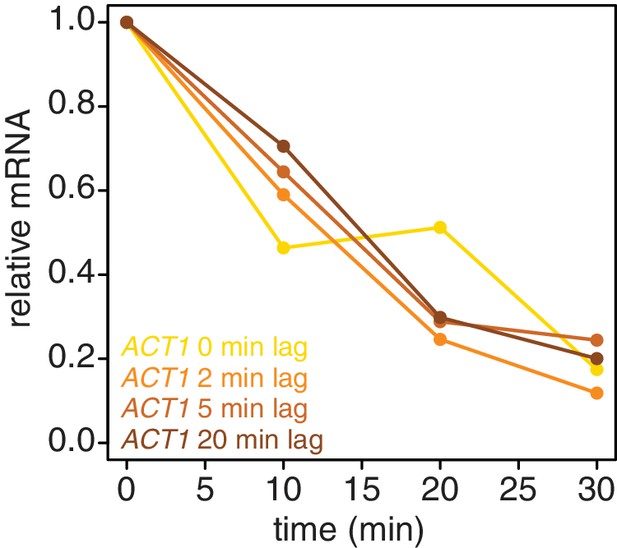
Introducing a brief lag period between the 4TU-chase and starting the decay timecourse improves the quality of the decay data.
Wild-type cells (KWY165) were subjected to the 4TU-chase protocol. A variable amount of time was allowed to elapse between the 4TU-chase and collection of the t = 0 sample and the unlabeled ACT1 mRNA levels were quantified.
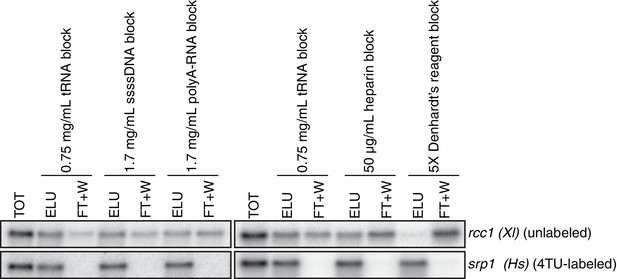
Analysis of streptavidin bead blocking efficiency.
Wild-type cells (KWY165) were labeled with 1 mM 4TU for 2 hr. 5 ng of in vitro transcribed rcc1 (Xl) mRNA spike and 5 ng of in vitro transcribed 4TU-labeled srp1α (Hs) mRNA spike were added to 10 μg of extracted total RNA and the RNA mix was biotinylated. mRNAs were enriched for using oligo-dT beads and the mRNAs were then subjected to streptavidin bead selection. Streptavidin beads were prepared using the indicated blocking agents and the total (TOT), eluate (ELU) and flowthrough plus washes (FT + W) fractions were analyzed by northern blot.
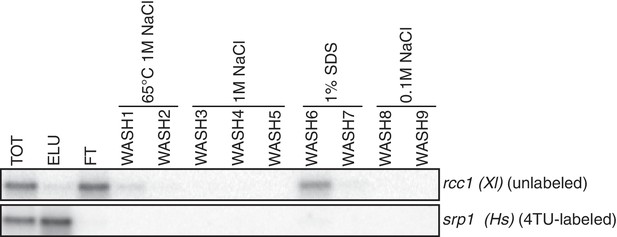
Analysis of unlabeled mRNA release during streptavidin bead purification.
RNA mixtures were prepared as in Appendix1—figure 7 and total (TOT), eluate (ELU) flowthrough (FT) and wash (WASH1-WASH9) samples were analyzed by northern blot.
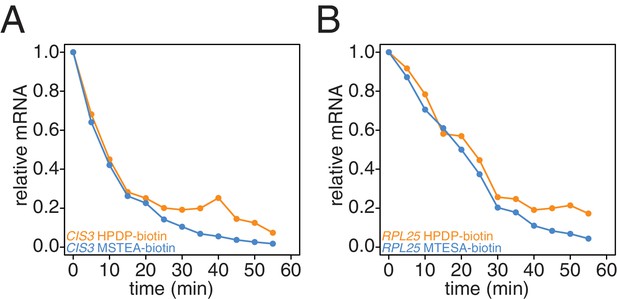
Comparison of HPDP-biotin with MTSEA-biotin in the efficiency of subtracting newly synthesized mRNAs during the chase phase.
Wild-type cells (KWY165) were subjected to the 4TU-chase protocol and RNAs were biotinylated with either HPDP-biotin (orange) or MTSEA-biotin (blue). Levels of unlabeled CIS3 and RPL25 mRNAs were determined.
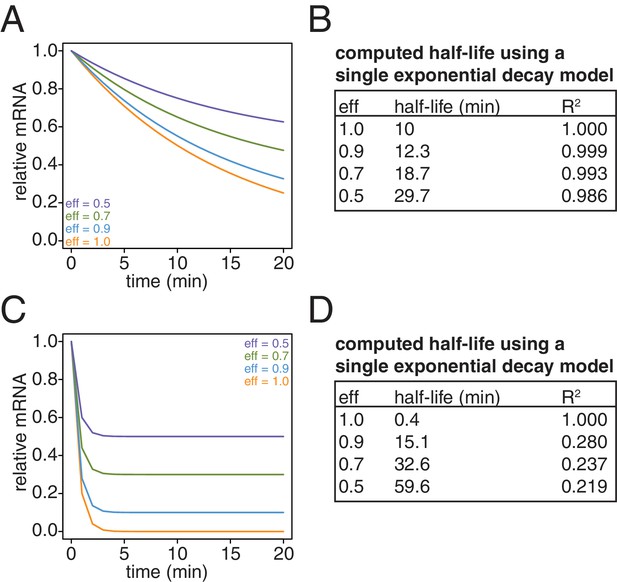
Simulation of the effects of non-ideal efficiencies in labeling and streptavidin bead separation on decay kinetics.
(A) Decay data for an mRNA with a 10 min half-life were simulated with variable degrees of labeling and purification efficiencies. (B) The simulated data in (A) were fit to a single exponential decay model (RNA(t)=RNA(0)*2 t/hl where hl is the half-life) and the half-lives and goodness of fits (R2) were determined. (C) Decay data for an mRNA with a 0.4 min half-life were simulated with variable degrees of labeling and purification efficiencies. (D) The simulated data in (C) were fit to a single exponential decay model (RNA(t)=RNA(0)*2 t/hl where hl is the half-life) and the half-lives and goodness of fits (R2) were determined.
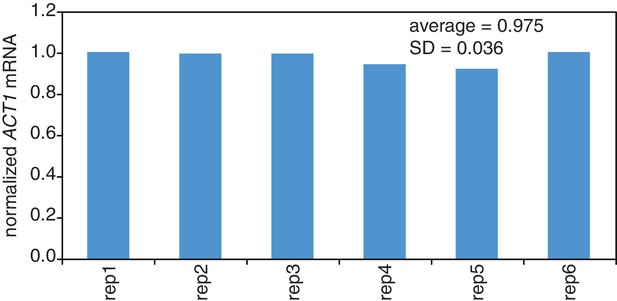
Reproducibility of cell lysis during RNA extraction.
Wild-type cells (KWY165) were grown to exponential phase and six technical replicate cell pellets of 5 OD600 units were collected. 10 ng of in vitro transcribed rcc1 (Xl) mRNA spike was added to each sample and the cells were subjected to the RNA extraction protocol. Levels of ACT1 and the spike mRNAs were determined and the ratio of ACT1:spike normalized to replicate one is plotted.
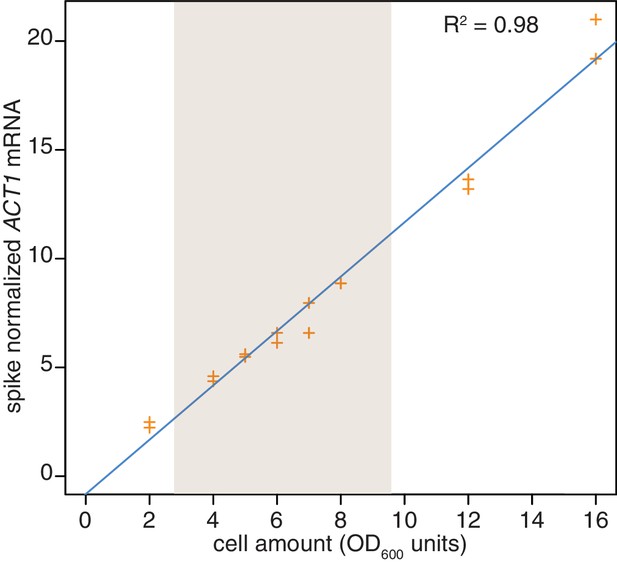
Reproducibility of cell lysis during RNA extraction.
Wild-type cells (KWY165) were grown to exponential phase and two technical replicate cell pellets of varying OD600 units were collected. 10 ng of in vitro transcribed rcc1 (Xl) mRNA spike was added to each sample and the cells were subjected to the RNA extraction protocol. Levels of ACT1 and the spike mRNAs were determined and the ratio of ACT1:spike is plotted. The shaded area indicates the OD600 range in which the 4TU-chase experiments are performed.
Additional files
-
Supplementary file 1
Sources of data for Figure 2.
- https://doi.org/10.7554/eLife.32536.022
-
Supplementary file 2
Yeast strains.
- https://doi.org/10.7554/eLife.32536.023
-
Supplementary file 3
Plasmids.
- https://doi.org/10.7554/eLife.32536.024
-
Supplementary file 4
qPCR primers.
- https://doi.org/10.7554/eLife.32536.025
-
Supplementary file 5
Plasmid sequences.
- https://doi.org/10.7554/eLife.32536.026
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32536.027





