Geometry of antiparallel microtubule bundles regulates relative sliding and stalling by PRC1 and Kif4A
Figures
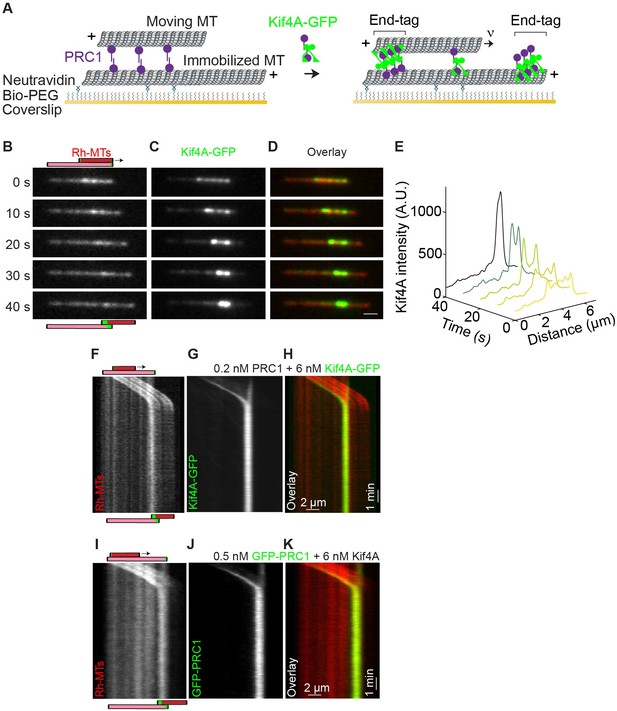
Relative microtubule sliding and the formation of stable antiparallel microtubule overlaps by PRC1 and Kif4A.
(A) Schematic of the in vitro assay. A biotinylated microtubule (‘immobilized MT’, X-rhodamine labeled) immobilized on a PEG coated coverslip and a non-biotinylated microtubule (‘moving MT’, X-rhodamine-labeled) are crosslinked in an antiparallel orientation by PRC1 (purple). Microtubule sliding and end-tag formation are initiated by addition of Kif4A-GFP (green), PRC1 and ATP. (B–D) Representative time-lapse fluorescence micrographs of relative microtubule sliding in experiments with 0.2 nM PRC1 and 6 nM Kif4A-GFP. Images show (B) a pair of X-rhodamine-labeled microtubules, (C) Kif4A-GFP, and (D) overlay images (red, microtubules; green, Kif4A-GFP). The schematic in (B) illustrates the position and relative orientation of both the immobilized (pink) and moving (red) microtubules and the end-tags (green) at the beginning and end of the time sequence. Scale bar: x: 2 µm. (E) Line scan analysis of the Kif4A intensity from the micrographs in (C) shows the distribution of Kif4A within the overlap at the indicated time points. (F–H) Kymographs show the relative sliding and stalling of antiparallel microtubules (F), associated Kif4A-GFP (G) and the overlay image (red, microtubules; green, Kif4A-GFP) (H). Assay condition: 0.2 nM PRC1 and 6 nM Kif4A-GFP. Scale bar: x: 2 µm and y: 1 min. (I–K) Kymographs show the relative sliding and stalling of antiparallel microtubules (I), associated GFP-PRC1 (J) and the overlay image (red, microtubules; green, GFP-PRC1) (K). Assay condition: 0.5 nM GFP-PRC1 and 6 nM Kif4A. Scale bar: x: 2 µm and y: 1 min.
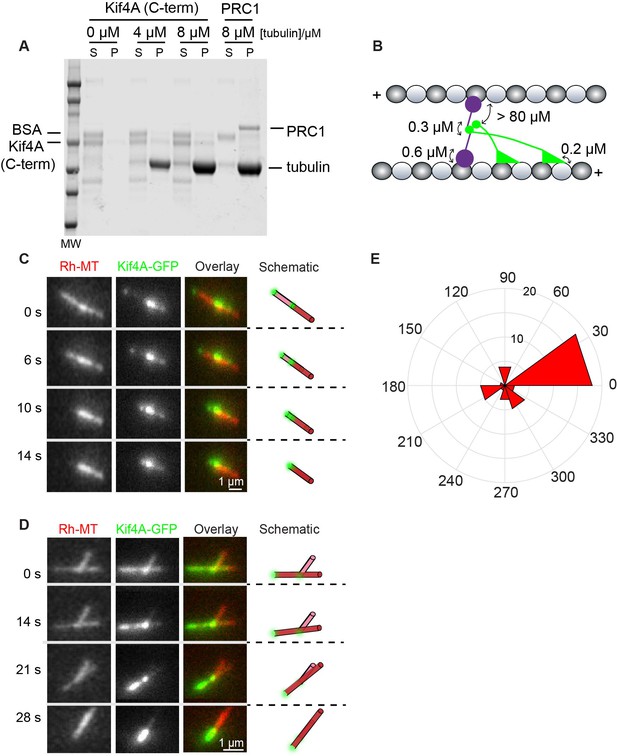
Molecular determinants of cross-bridging and sliding in the PRC1-Kif4A system.
(A) Microtubule co-sedimentation assay. SDS Page analysis of the interaction of Kif4A (C-term) with increasing amounts microtubules (0–8 μM). Full-length PRC1 was included as a control. Quantification of band intensities: Kif4A_pellet = 8% and 6% at 4 μM and 8 μM tubulin. BSA_pellet = 1% and 3% at 4 μM and 8 μM tubulin. (B) Schematic shows the proposed molecular configuration of the cross-bridging PRC1-Kif4A complex in a microtubule overlap. Known dissociation constants are indicated. (C–D) Representative time-lapse fluorescence micrographs of relative microtubule sliding in experiments with 6 nM Kif4A-GFP + 2 mM ATP. Images show a pair of X-rhodamine-labeled microtubules, Kif4A-GFP, and overlay images (red, microtubules; green, Kif4A-GFP). The schematic illustrates the position and relative orientation of both the immobilized and moving microtubules (red) and the end-tags (green) at the beginning and end of the time sequence. (E) Rose diagram of the initial angle of attachment of the sliding microtubule. The plot shows the most probable angle of attachment is between 0–30° (N = 39). Assay condition: 6 nM Kif4A-GFP + 2 mM ATP.
-
Figure 2—source data 1
This spreadsheet contains the initial angle of attachment of the sliding microtubule used to generate the rose diagram shown in Figure 2E.
- https://doi.org/10.7554/eLife.32595.004
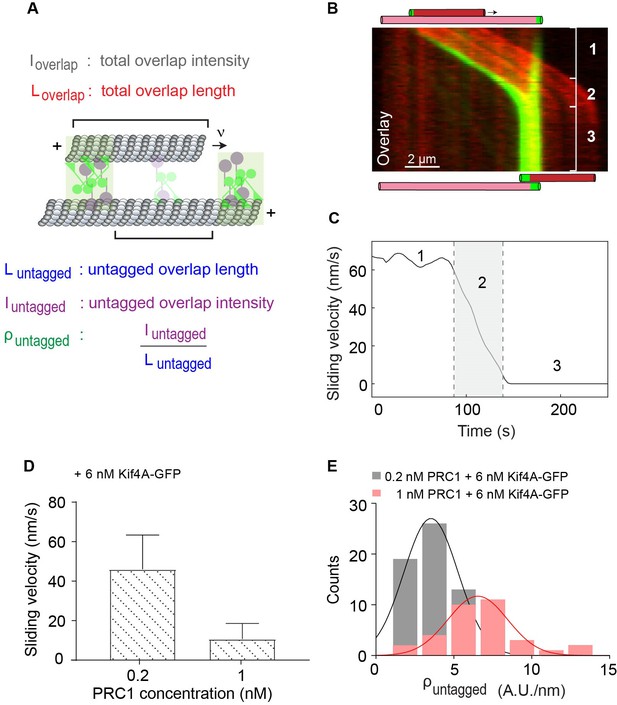
Quantitative analysis of microtubule sliding in the PRC1-Kif4A system.
(A) Schematic of a pair of crosslinked microtubules showing the parameters described in Figures 1 and 4. (B) Kymograph shows the relative sliding and stalling in a pair of antiparallel microtubules (red) and associated Kif4A-GFP (green). Assay condition: 0.2 nM PRC1 and 6 nM Kif4A-GFP. Scale bar: 2 µm. The schematic illustrates the position and relative orientation of both the immobilized (pink) and moving (red) microtubules and the end-tags (green) at the beginning and end of the time sequence. (C) Time record of the instantaneous sliding velocity of the moving microtubule derived from the kymograph in (B). The dashed lines demarcate the three phases observed in the sliding velocity profile: (1) constant phase, (2) slow down and (3) stalling. (D) Bar graph of the average sliding velocity calculated from the constant velocity movement in phase-1. Assay conditions: (i) 0.2 nM and 6 nM Kif4A-GFP (mean: 46 ± 17; N = 98) (ii) 1 nM PRC1 and 6 nM Kif4A-GFP (mean: 11 ± 8; N = 45). Error bars represent the standard deviation of the data. (E) Histograms of the initial GFP-fluorescence density in the untagged region of the overlap, . Assay conditions: (i) 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; mean: 3.5 ± 1.7 A.U./nm; N = 64) and (ii) 1 nM PRC1 and 6 nM Kif4A-GFP (red; mean: 6.5 ± 1.9 A.U./nm; N = 33). The mean and error values were obtained by fitting the histograms to a Gaussian distribution.
-
Figure 3—source data 1
This spreadsheet contains the average sliding velocity used to generate the bar graph shown in Figure 3D and the initial GFP-fluorescence density, , used to generate Figure 3E.
- https://doi.org/10.7554/eLife.32595.016
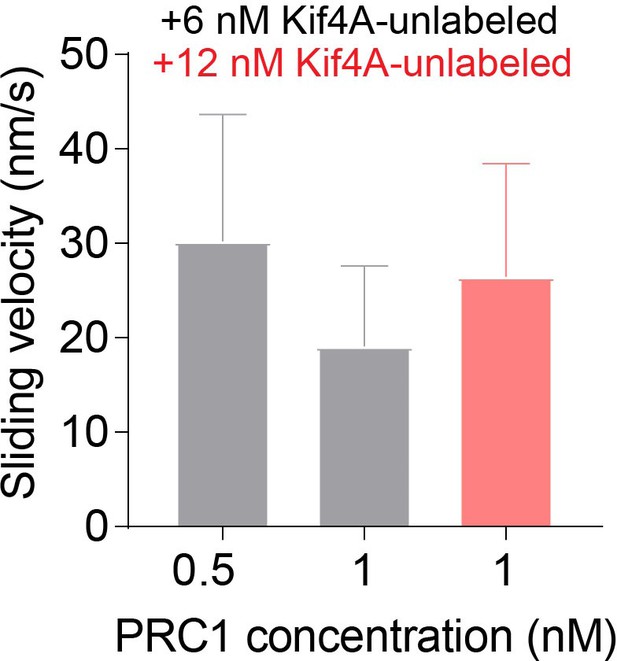
Average sliding velocity of antiparallel microtubule overlaps by PRC1 and Kif4A.
Bar graph of the average sliding velocity calculated from the constant velocity movement in phase-1. Assay conditions: (i) 0.5 nM GFP-PRC1 and 6 nM Kif4A (black; N = 33), (ii) 1 nM GFP-PRC1 with 6 nM Kif4A (black; N = 41) and (iii)1 nM GFP-PRC1 with 12 nM Kif4A (red; N = 46). Error bars represent the standard deviation of the data.
-
Figure 3—figure supplement 1—source data 1
This spreadsheet contains the average sliding velocity used to generate the bar graph.
- https://doi.org/10.7554/eLife.32595.017
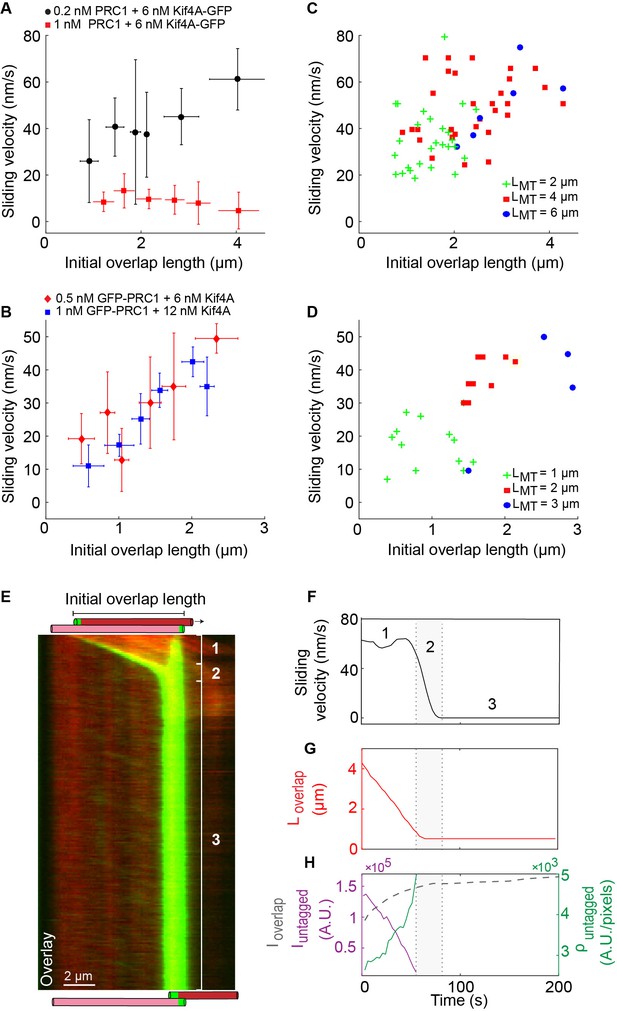
Microtubule sliding velocity in the PRC1-Kif4A system scales with initial overlap width.
(A–B) Binned plots of initial sliding velocity versus the initial overlap length. The initial overlap length between the moving MT and immobilized MT is calculated from the rhodamine MT channel. Sliding velocity is calculated from the constant velocity movement in phase-1. (A) Assay conditions: (i) 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 60; Pearson’s correlation coefficient = 0.54) and (i) 1 nM PRC1 and 6 nM Kif4A-GFP (red; N = 42; Pearson’s correlation coefficient = 0.03). (B) Assay conditions: (i) 0.5 nM GFP-PRC1 and 6 nM Kif4A (red; N = 25; Pearson’s correlation coefficient = 0.69) and (ii) 1 nM GFP-PRC1 and 12 nM Kif4A (blue; N = 20; Pearson’s correlation coefficient = 0.74). (C–D) Scatter plot of the average sliding velocity versus the initial overlap length color-coded by moving microtubule length, . (C) Assay condition: 0.2 nM PRC1 and 6 nM Kif4A-GFP (green: = 2 ± 0.5 µM, red: = 4 ± 0.5 µM, blue: = 6 ± 0.5 µM; N = 60). (D) Assay condition: 0.5 nM GFP-PRC1 and 6 nM Kif4A (green: = 1 ± 0.5 µM, red: = 2 ± 0.5 µM, blue: = 3 ± 0.5 µM; N = 25). (E) Kymograph shows the relative sliding and stalling in a pair of antiparallel microtubules (red) and associated Kif4A-GFP (green). Assay condition: 0.2 nM PRC1 and 6 nM Kif4A-GFP. Scale bar: 2 µm. The schematic illustrates the position and relative orientation of both the immobilized (pink) and moving (red) microtubules and the end-tags (green) at the beginning and end of the time sequence. (F) Time record of the instantaneous sliding velocity of the moving microtubule derived from the kymograph in (E). The dashed lines demarcate the three phases observed in the sliding velocity profile: (1) constant phase, (2) slow down and (3) stalling. (G) Time record of the overlap length (red; ) derived from the kymograph in (E). (H) Time record of the total fluorescence intensity in the antiparallel overlap (dashed gray; ), fluorescence intensity in the untagged region of the overlap (solid purple; ), and fluorescence density (intensity per unit overlap length) in the untagged region of the overlap (solid green; ) derived from the kymograph in (E).
-
Figure 4—source data 1
This spreadsheet contains the sliding velocity versus the initial overlap length used to generate the binned plots shown in Figure 4A–B and the scatter plots shown in Figure 4C–D.
- https://doi.org/10.7554/eLife.32595.011
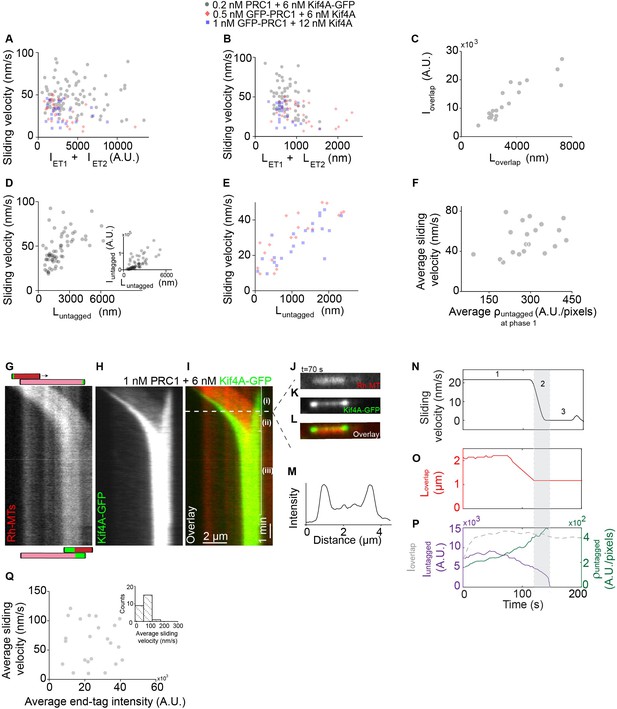
Relative microtubule sliding and the formation of stable antiparallel microtubule overlaps by PRC1 and Kif4A.
(A) Scatter plot of the average sliding velocity versus the sum of end-tag intensity, . Assay conditions: 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 95; Pearson’s correlation coefficient = −0.18) and (ii) 0.5 nM GFP-PRC1 and 6 nM Kif4A (red; N = 32; Pearson’s correlation coefficient = −0.37), and (iii) 1 nM GFP-PRC1 with 12 nM Kif4A (blue; N = 27; Pearson’s correlation coefficient = −0.44). (B) Scatter plot of the average sliding velocity versus the sum of end-tag length, . Assay conditions: (i) 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 66; Pearson’s correlation coefficient= −0.35) and (ii) 0.5 nM GFP-PRC1 and 6 nM Kif4A (red; N = 29; Pearson’s correlation coefficient = −0.40), and (iii)1 nM GFP-PRC1 with 12 nM Kif4A (blue; N = 26; Pearson’s correlation coefficient = −0.40). (C) Scatter plot of the overlap intensity, , versus the overlap length, (N = 20; Pearson’s correlation coefficient = 0.90). (D) Scatter plot of the average sliding velocity versus the untagged overlap length, . Assay conditions: 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 64; Pearson’s correlation coefficient = 0.54). The inset shows the untagged overlap intensity, , versus the untagged overlap length, (Pearson’s correlation coefficient = 0.55). (E) Scatter plot of the average sliding velocity versus the untagged overlap length, . Assay conditions: (i) 0.5 nM GFP-PRC1 and 6 nM Kif4A (red; N = 20; Pearson’s correlation coefficient = 0.83), and (ii)1 nM GFP-PRC1 with 12 nM Kif4A (blue; N = 22; Pearson’s correlation coefficient = 0.79). (F) Scatter plot of the average sliding velocity versus average untagged density () at phase-1. Assay conditions: 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 20; Pearson’s correlation coefficient = -0.15). (G–I) Kymographs show the relative sliding and stalling of antiparallel microtubules (G), associated Kif4A-GFP (H) and the overlay image (red, microtubules; green, Kif4A-GFP) (I). Assay condition: 1 nM PRC1 and 6 nM Kif4A-GFP. The schematic in (G) illustrates microtubule orientations and positions at the start and end of the experiment (red, microtubules; green, plus-end tag). Scale bar: x: 2 µm and y: 1 min. (J–L) Fluorescence micrographs from the microtubule pair in (I) at = 70 s. Images show (J) a pair of X-rhodamine-labeled microtubules, (K) Kif4A-GFP, and (L) overlay images (red, microtubules; green, Kif4A-GFP). (M) Line scan analysis of the Kif4A intensity from the micrograph in (K) shows the distribution of Kif4A within the overlap at the indicated time point. (N) Time record of the instantaneous sliding velocity of the moving microtubule derived from the kymograph in (G). The dashed lines demarcate the three phases observed in the sliding velocity profile: (i) constant phase, (ii) slow down and (iii) stalling. (O) Time record of the total overlap length (red; ) derived from the kymograph in (G). (P) Total fluorescence intensity (dashed gray; ), total fluorescence intensity in the untagged region of the overlap (solid purple; ), and fluorescence density in the untagged region of the overlap (solid green; ) derived from the kymograph in (H). (Q) Scatter plot of the average sliding velocity versus end-tag intensity of Kif4A-GFP alone from experiments in Figure 2. The inset shows the histogram of the average sliding velocity (mean: 75 ± 25 nm/s; N = 25). Assay condition: 6 nM Kif4A-GFP + 2 mM ATP.
-
Figure 4—figure supplement 1—source data 1
This spreadsheet contains (1) the average sliding velocity versus the sum of end-tag intensity, , used to generate the scatter plot in Figure 4—figure supplement 1A, (2) the average sliding velocity versus the sum of end-tag length, , used to generate the scatter plot in Figure 4—figure supplement 1B, (3) the overlap intensity, , versus the overlap length, used to generate the scatter plot in Figure 4—figure supplement 1C, (4) the average sliding velocity versus the untagged overlap length, , used to generate the scatter plots in Figure 4—figure supplement 1D-E, (5) the average sliding velocity versus average untagged density () at phase-1 used to generate the scatter plot in Figure 4—figure supplement 1F and (6) the average sliding velocity versus end-tag intensity of Kif4A-GFP used to generate the scatter plot and histogram in Figure 4—figure supplement 1Q.
- https://doi.org/10.7554/eLife.32595.012
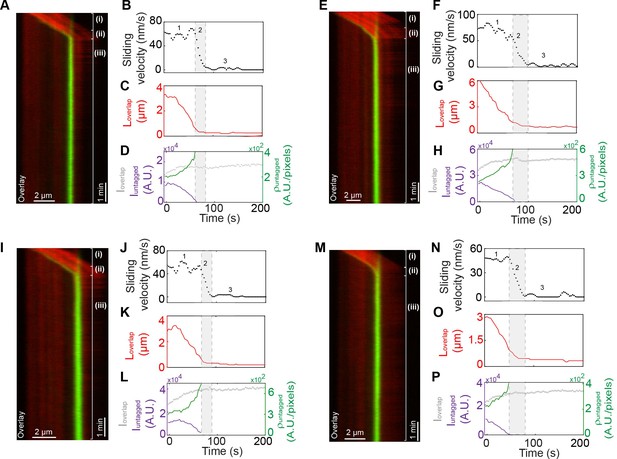
Kymographs and the corresponding quantification of microtubule sliding and stopping events.
(A–P) (A) Kymograph shows the relative sliding and stalling in a pair of antiparallel microtubules (red) and associated Kif4A-GFP (green). Assay condition: 0.2 nM PRC1 and 6 nM Kif4A-GFP. Scale bar: 2 µm. (B) Time record of the instantaneous sliding velocity of the moving microtubule derived from the kymograph in (A). The dashed lines demarcate the three phases observed in the sliding velocity profile: (1) constant phase, (2) slow down and (3) stalling. (C) Time record of the overlap length (red; ) derived from the kymograph in (A). (D) Time record of the total fluorescence intensity in the antiparallel overlap (dashed gray; ), fluorescence intensity in the untagged region of the overlap (solid purple; ), and fluorescence density (intensity per unit overlap length) in the untagged region of the overlap (solid green; ) derived from the kymograph in (A). (E–H, I–L, M–P) Same as (A–D).
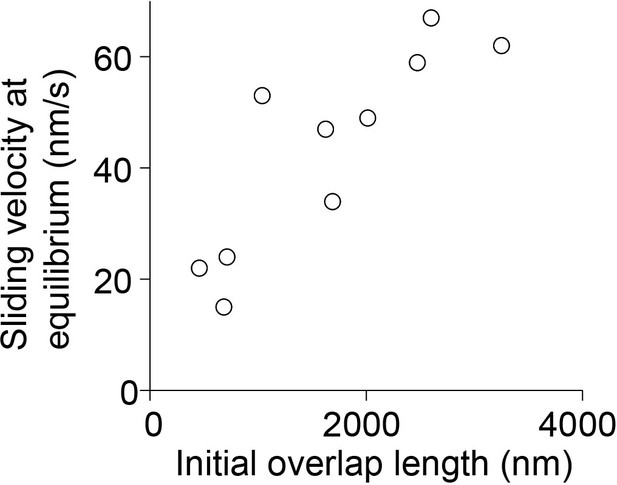
Analysis of sliding velocity and initial overlap length at equilibrium.
Scatter plot of the sliding velocity as a function of initial overlap length at equilibrium condition where reaches a constant value in the microtubule overlap. Assay conditions: 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 10).
-
Figure 4—figure supplement 3—source data 2
This spreadsheet contains the sliding velocity as a function of initial overlap length at equilibrium used to generate the scatter plot.
- https://doi.org/10.7554/eLife.32595.013
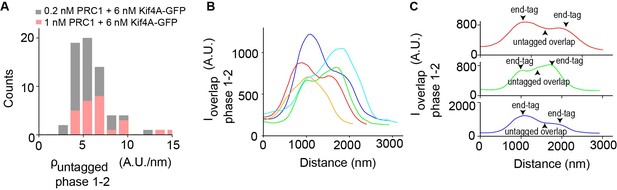
Protein density at phase 1–2 transition in microtubule overlaps.
(A) Histogram of the untagged density , in the microtubule overlap at phase 1-2 transition. Assay conditions: (i) 0.2 nM PRC1 + 6 nM Kif4A-GFP (black; mean: 5 A.U./nm; N = 64) and (ii) 1 nM PRC1 + 6 nM Kif4A-GFP (red; mean: 7 A.U./nm; N = 26). (B) Line scan analysis of the Kif4A intensity from five kymographs which show the distribution of Kif4A within the overlap at the phase 1–2 transition. (C) Zoomed-in view of three events in (B) showing end-tag and untagged overlap regions.
-
Figure 4—figure supplement 4—source data 3
This spreadsheet contains untagged density , in the microtubule overlap at phase 1-2 transition used to generate the histogram in Figure 4—figure supplement 4A.
- https://doi.org/10.7554/eLife.32595.014
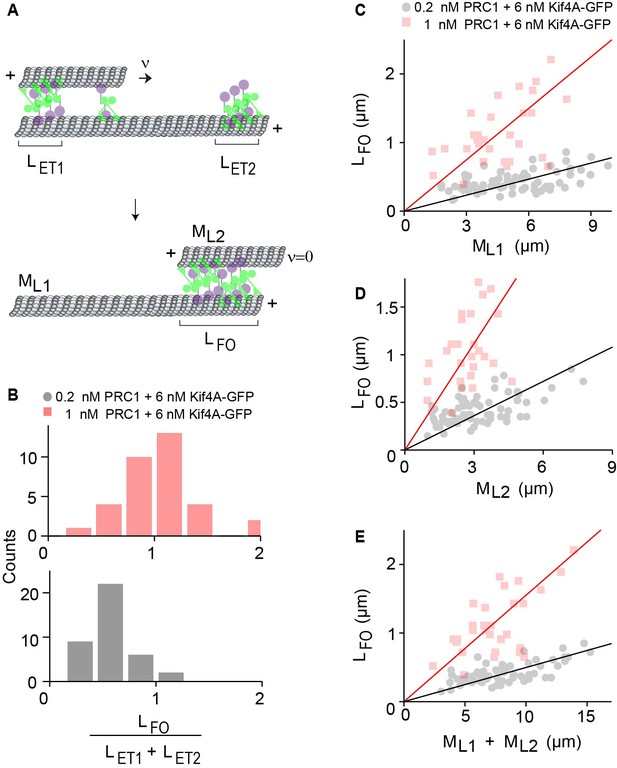
The width of the final antiparallel overlap established by PRC1 and Kif4A is determined by end-tag and microtubule lengths.
(A) Schematic shows the formation of a stable antiparallel overlap upon collision of the two end-tags and the stalling of relative microtubule sliding. The initial overlap length is the overlap length of the moving MT on the immobilized MT at = 0. and are the lengths of the end-tags consisting Kif4A and PRC1 on the plus-end of each MT. The moving MT with length moves relative to the immobilized MT with length , at velocity = . The collision and the stalling of the end-tags form a stable overlap, which is the final overlap length at = 0. (B) Histograms of the ratio of sum of the end-tag lengths () and final overlap length . Assay conditions: (i) 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 39) and (ii) 1 nM PRC1 and 6 nM Kif4A-GFP (red; N = 33). (C–E) Plots of the final overlap length () versus (C) the immobilized microtubule length (), (D) moving microtubule length (), and (E) and the sum of microtubule lengths (). Assay conditions: (i) 0.2 nM PRC1 and 6 nM Kif4A-GFP (black; N = 68) and (ii) 1 nM PRC1 and 6 nM Kif4A-GFP (red; N = 30). The Pearson’s correlation coefficient for (E) is (i) 0.65 and (ii) 0.62.
-
Figure 5—source data 1
This spreadsheet contains the ratio of the sum of the end-tag lengths () and final overlap length ( used to generate the histogram in Figure 5B and the final overlap length () versus the immobilized microtubule length (), moving microtubule length (), and the sum of microtubule lengths () used to generate the plots in Figure 5C-E.
- https://doi.org/10.7554/eLife.32595.020
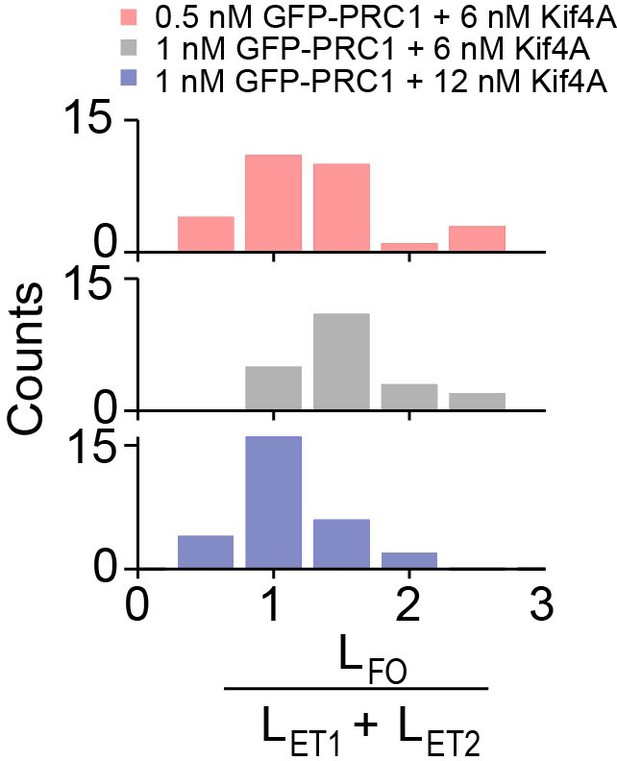
The width of the final antiparallel overlap established by PRC1 and Kif4A is determined by end-tag and microtubule lengths.
Histograms of the ratio of sum of the end-tag lengths () and final overlap length (). Assay conditions: (i) 0.5 nM PRC1 and 6 nM Kif4A-GFP (red; mean = 0.7 ± 0.4; N = 29) and (ii) 1 nM PRC1 and 6 nM Kif4A-GFP (black; mean = 1.2 ± 0.3; N = 21), (iii) 1 nM PRC1 and 12 nM Kif4A-GFP (blue; mean=0.7 ± 0.4; N = 28).
-
Figure 5—figure supplement 1—source data 1
This spreadsheet contains the ratio of sum of the end-tag lengths () and final overlap length () used to generate the histogram.
- https://doi.org/10.7554/eLife.32595.021
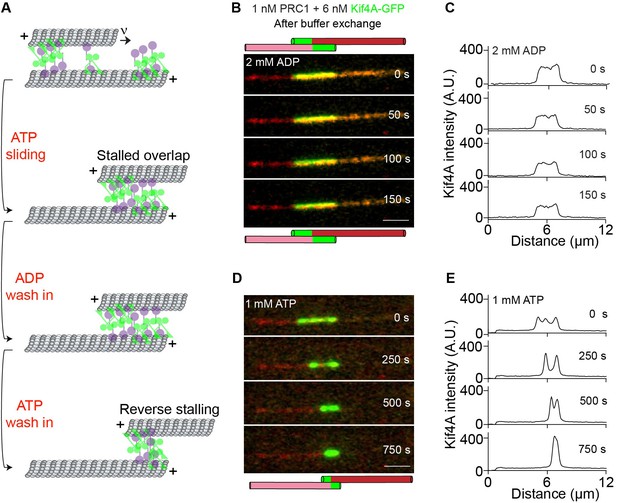
Examination of the mechanisms that ensure stability of the overlaps established by PRC1 and Kif4A.
(A) Schematic of the ADP and ATP wash-in experiments performed with stalled microtubule overlaps Figure 6B-E. (B–E) The following figures are representative dual-channel fluorescence micrographs showing microtubules (red) and associated Kif4A-GFP (green) under different experimental conditions. (B–C) Time-lapse images (B) and corresponding line-scan profiles (C) of Kif4A-GFP fluorescence of a microtubule pair established as in (Figure 1A ) and subsequent exchange into a buffer containing 2 mM ADP. (D–E) Time-lapse images (D) and corresponding line-scan profiles (E) of Kif4A-GFP fluorescence of the microtubule pair in (D) after flowing in 1 mM ATP into the chamber. Scale bar: 2 µm.
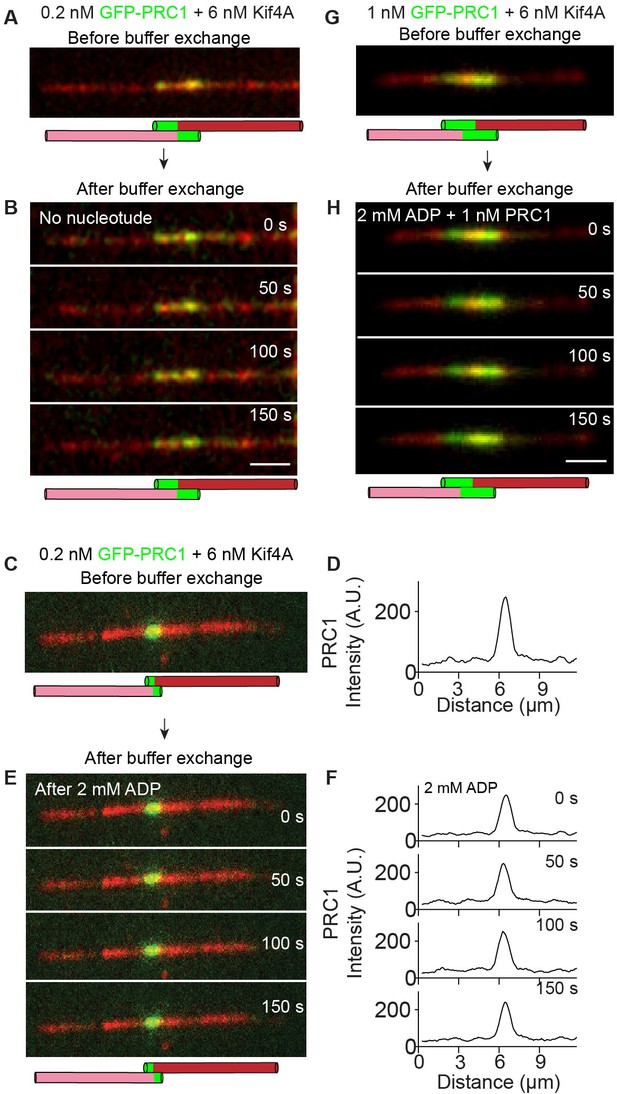
Examination of the mechanisms that maintain the stable antiparallel overlaps established by PRC1 and Kif4A.
Representative dual-channel fluorescence micrographs showing microtubules (red) and associated GFP-PRC1 (green). (A) Image shows a stable steady-state antiparallel microtubule overlap established in the presence of 0.2 nM GFP-PRC1, 6 nM Kif4A and 1 mM ATP. (B) Time-lapse images of the same microtubule pair shown in (A) after flowing in buffer containing no nucleotide into the chamber. (C–D) Time lapse image (C) and corresponding line-scan profile (D) show a stable steady-state antiparallel microtubule overlap established in the presence of 0.2 nM GFP-PRC1, 6 nM Kif4A and 1 mM ATP. (E–F) Time-lapse images (E) and corresponding line-scan profiles (F) of GFP-PRC1 fluorescence of the microtubule pair in (E) after flowing in buffer containing 2 mM ADP. (G) Image shows a stable steady-state antiparallel microtubule overlap established in the presence of 1 nM GFP-PRC1, 6 nM Kif4A and 1 mM ATP. (H) Time-lapse images of the same microtubule pair shown in (G) after flowing in 2 mM ADP and 1 nM PRC1 into the chamber. Scale bar: 2 µm.
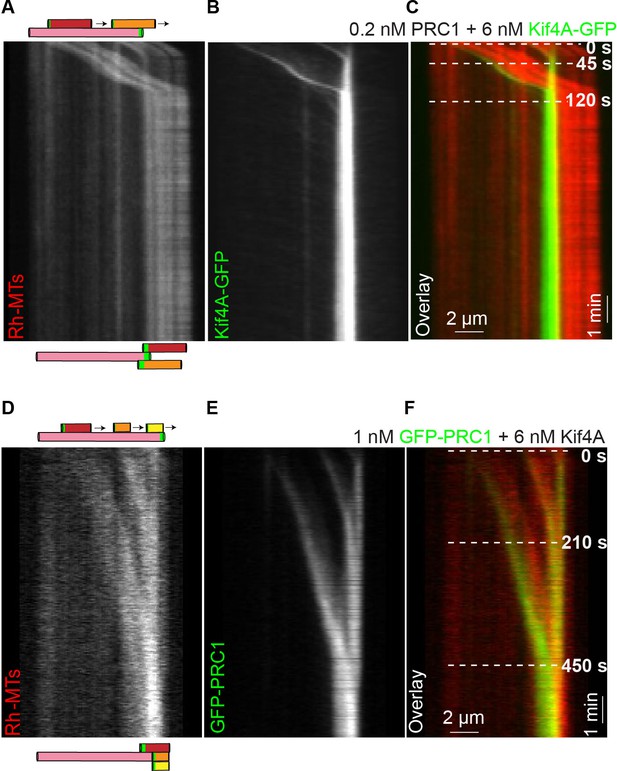
Antiparallel array composed of multiple microtubules are aligned at microtubule plus-ends formed by PRC1 and Kif4A.
(A–C) Kymographs show the relative sliding of two microtubules relative to an immobilized microtubule (A), associated Kif4A-GFP (B) and the overlay image (red, microtubules; green, Kif4A-GFP) (C). Both moving microtubules stall at the plus-end of the immobilized microtubule. Assay condition: 0.2 nM PRC1 and 6 nM Kif4A-GFP. Scale bar: x: 2 µm and y: 1 min. (D–F) Kymographs show the relative sliding of three microtubules relative to an immobilized microtubule (D), associated GFP-PRC1 (E) and the overlay image (red, microtubules; green, GFP-PRC1) (F). All three moving microtubules stall at the plus-end of the immobilized microtubule. Assay condition: 1 nM GFP-PRC1 and 6 nM Kif4A. Scale bar: x: 2 µm and y: 1 min.
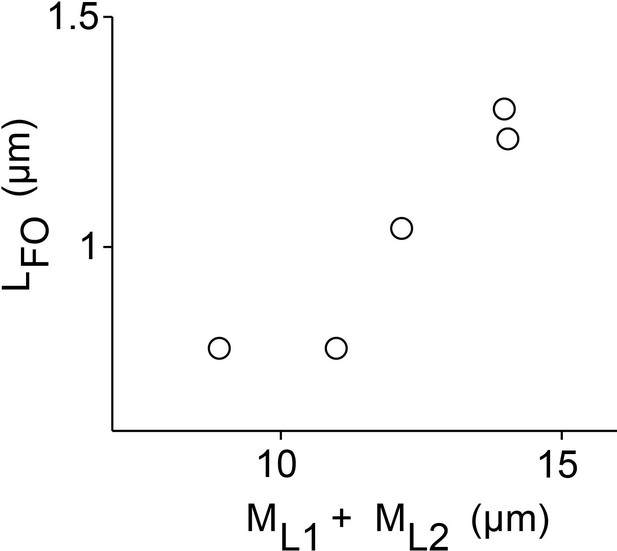
Multiple microtubule sliding by PRC1 and Kif4A.
Plot of the final overlap length () versus the sum of microtubule lengths () (N = 5) for multiple microtubule sliding as in Figure 7.
-
Figure 7—figure supplement 1—source data 1
This spreadsheet contains the final overlap length () versus the sum of microtubule lengths () used to generate the plot.
- https://doi.org/10.7554/eLife.32595.026
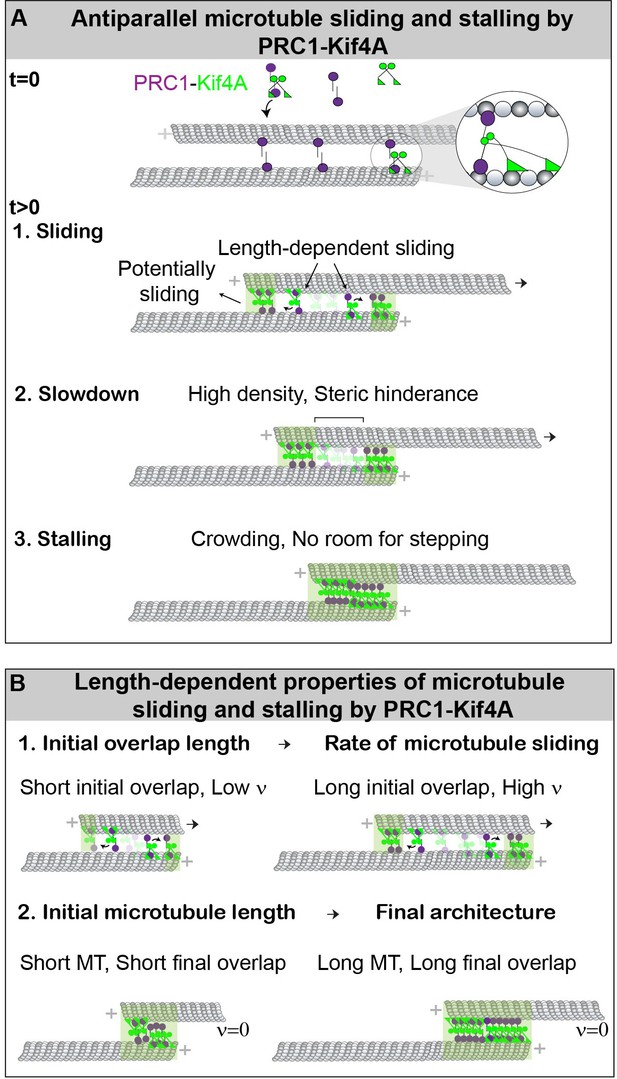
Model for the length-dependent sliding by the collective activity of PRC1 and Kif4A.
(A) Mechanism of microtubule sliding and stalling by PRC1 and Kif4A. At the initial state, = 0, the ‘immobilized’ and ‘moving’ microtubules are crosslinked by PRC1 to form an antiparallel overlap. The zoomed-in view shows the proposed molecular configuration of the cross-bridging PRC1-Kif4A complex in a microtubule overlap. At > 0, Kif4A molecules are introduced into the solution, which form a complex with PRC1. This initiates the formation of PRC1-Kif4A end-tags at the plus-ends of both microtubules as well as relative sliding of the moving microtubule. The sliding of microtubules is most likely due to the cross-bridging molecules in the untagged overlap. The slowdown is likely due to the high density of molecules and steric hindrance to motor stepping when the end-tags arrive at close proximity. This eventually halts movement when the end-tags merge, and a stable overlap is established. (B) The schematic shows the length-dependent properties of initial microtubule sliding and subsequent stalling of overlapping antiparallel microtubules established by PRC1 and Kif4A. Our experiments show that: (1) Microtubules that form shorter initial overlaps slide with lower velocity than microtubule pairs that form longer initial overlaps. (2) Since the size of PRC-Kif4A end-tags scale with microtubule length, shorter microtubules form a short overlap and the longer microtubules form a long overlap.
-
Figure 8—source data 1
This table contains the sliding efficiencies calculated using Equation 3 for = 1-10%.
- https://doi.org/10.7554/eLife.32595.030
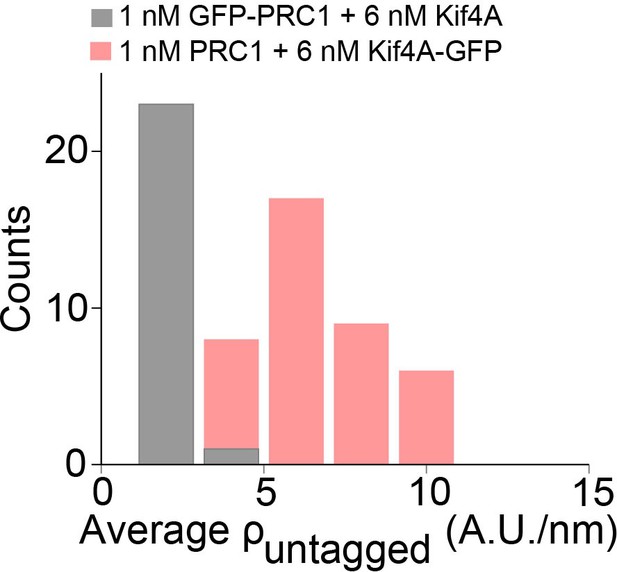
Average untagged protein density in microtubule overlaps.
Histogram of the average untagged density (). Assay conditions: (i) 1 nM GFP-PRC1 + 6 nM Kif4A (black; 2 ± 1 A.U./nm; N = 24) and (ii) 1 nM PRC1 + 6 nM Kif4A-GFP (red; mean: 6 ± 1 A.U./nm; N = 40).
-
Figure 8—figure supplement 1—source data 1
This spreadsheet contains the average untagged density () used to generate the histogram.
- https://doi.org/10.7554/eLife.32595.031
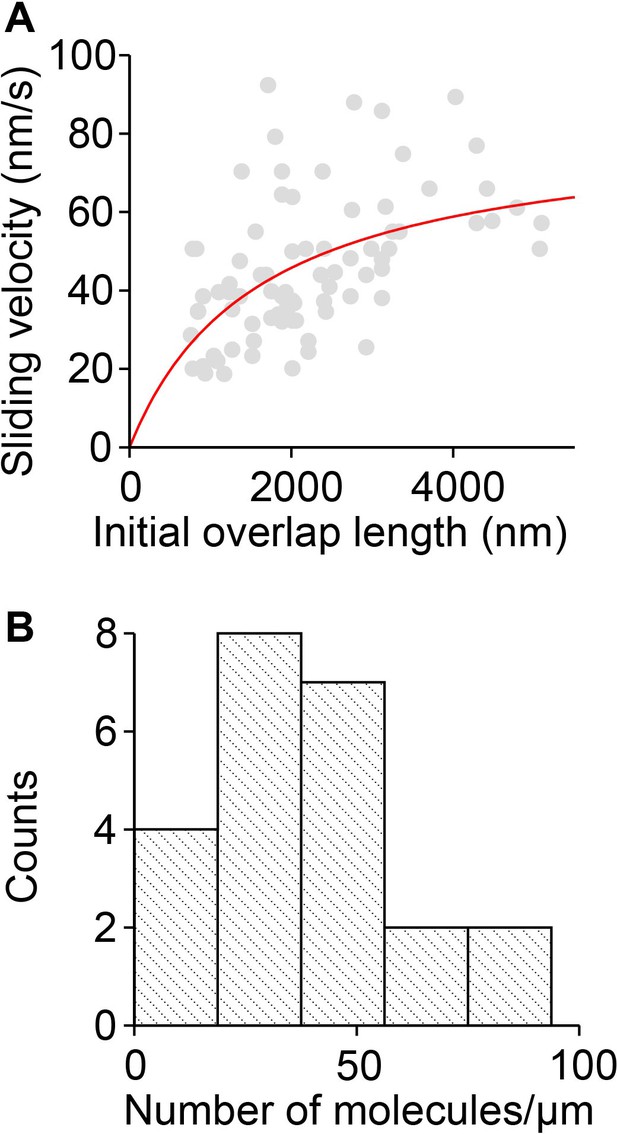
Estimation of the sliding efficiency of PRC1-Kif4A molecules.
(A) The fitting of Equation 1 (red line) to the microtubule sliding velocity as a function of initial microtubule overlap length data (Assay condition: 0.2 nM PRC1 + 6 nM Kif4A-GFP (gray circles; N = 83). (B) Estimation of the number of molecules/µm from experimental fluorescence intensity measurements.
-
Figure 8—figure supplement 2—source data 2
This spreadsheet contains the microtubule sliding velocity versus the initial microtubule overlap length data used to generate the scatter plot in Figure 8—figure supplement 2A and the estimation of the number of molecules/µm used to generate the histogram in Figure 8—figure supplement 2B.
- https://doi.org/10.7554/eLife.32595.032
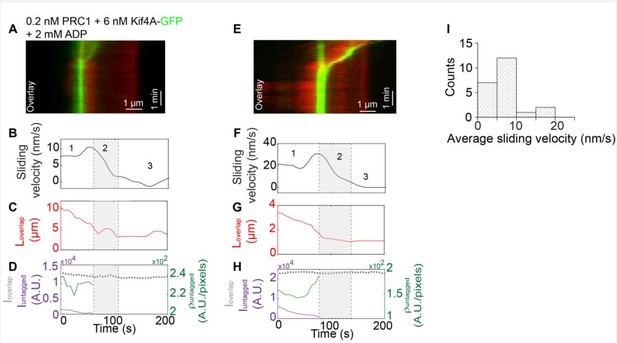
Method:Biotinylated rhodamine-labeled microtubules were immobilized in a flow chamber.
0.2 nM un-labeled PRC1 in assay buffer was flushed into the flow chamber. Next, non-biotinylated microtubules were flushed in the flow cell and incubated for 10-15 mins to allow antiparallel overlap formation. Afterwards, 6 nM Kif4A-GFP and 2 mM ADP were flowed into the chamber. To visualize microtubule sliding, 2 mM ATP were flowed into the chamber in assay buffer and a time-lapse sequence of images was immediately acquired at a rate of 5 frames/s for 10-15 min.
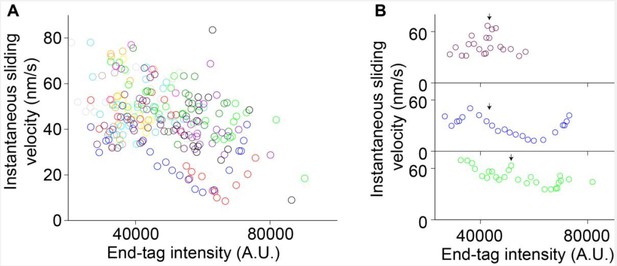
Instantaneous sliding velocity versus moving end-tag intensity, 𝐼𝐸𝑇2.
(A) Instantaneous sliding velocity versus moving end-tag intensity, 𝐼𝐸𝑇2, plots from 16 kymographs (colored circles). (B) Zoomed-in view of three events in (A). The arrows indicate the phase 1-2 transition point.
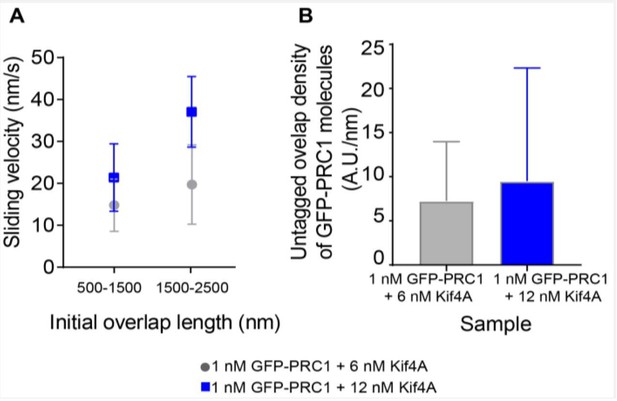
Sliding velocity as a function of initial overlap length and untagged overlap density.
A) Sliding velocity as a function of initial overlap length for two bin sizes. Assay condition: 1 nM GFP-PRC1 + 6 nM Kif4A (gray; 500-1500 nm: N = 13, 1500-2500 nm: N = 18) and 1 nM GFP-PRC1 + 12 nM Kif4A (blue; 500-1500 nm: N = 13, 1500-2500 nm: N = 9) B) Histogram of the untagged overlap density of GFP-PRC1. Assay condition: 1 nM GFP-PRC1 + 6 nM Kif4A (gray; N = 38) and 1 nM GFP-PRC1 + 12 nM Kif4A (blue; N = 18).
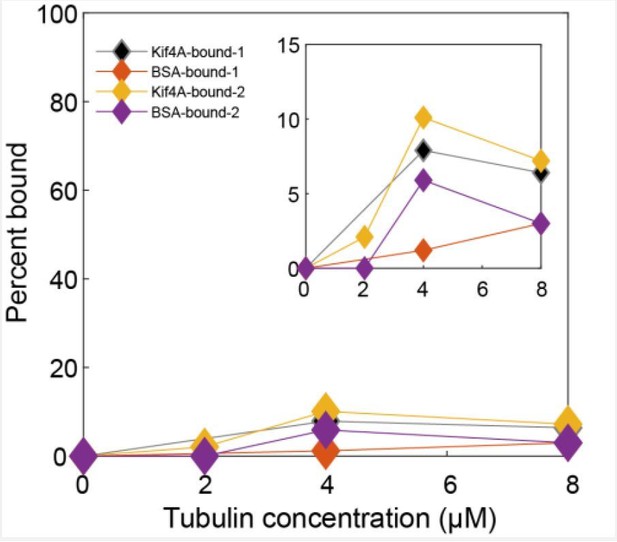
Quantitative analysis of SDS-PAGE gels from two co-sedimentation assays.
The percentage of Kif4A (black and orange) and BSA (red and purple) in the pellet is plotted against tubulin concentration. The curves labeled bound-1 are connected to the gel in Figure 2 of the main text. Inset: Zoomed-in view of the plot.
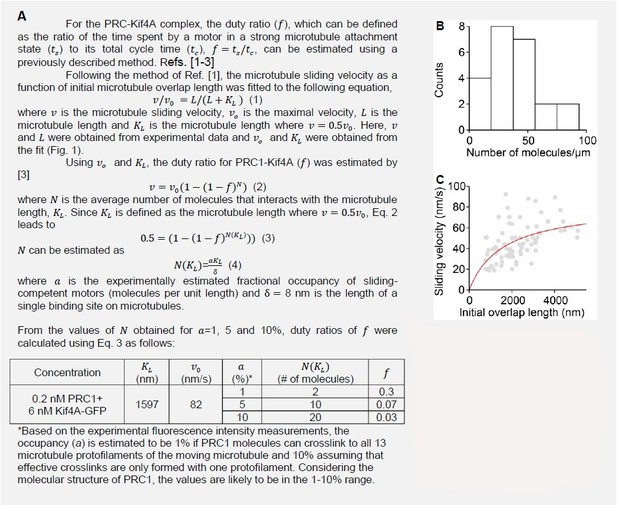
A) Calculation of the duty ratio of the PRC1-Kif4A molecules. B) Estimation of the number of molecules/µm from experimental fluorescence intensity measurements. C) The fitting of Eq. 1. (red line) to the microtubule sliding velocity as a function of initial microtubule overlap length data (Assay condition: 0.2 nMPRC1 + 6 nM Kif4A-GFP (gray circles; N = 84).
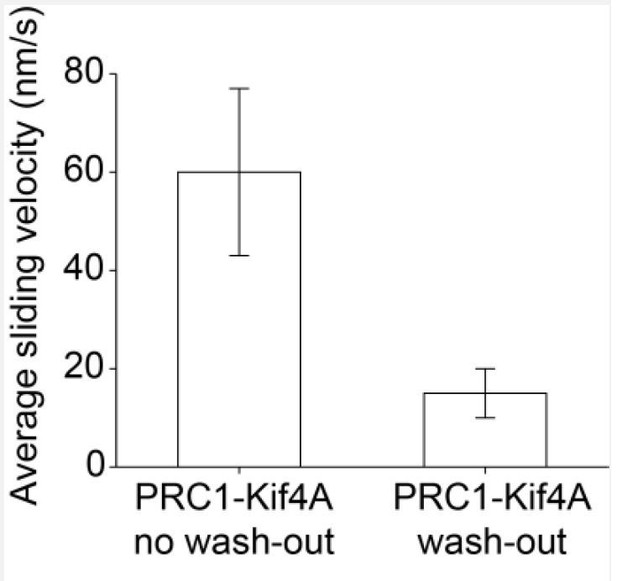
Average sliding velocity for PRC1-Kif4A no wash-out experiment (0.2 nM PRC1 + 6 nM Kif4A-GFP, N = 84; 60 ± 17nm/s) and wash-out experiment (0.2 nM PRC1 + 6 nM Kif4A-GFP + 2 mM ADP, N = 22; 15 ± 5 nm/s).
https://doi.org/10.7554/eLife.32595.040Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32595.033





