Semisynthetic biosensors for mapping cellular concentrations of nicotinamide adenine dinucleotides
Figures
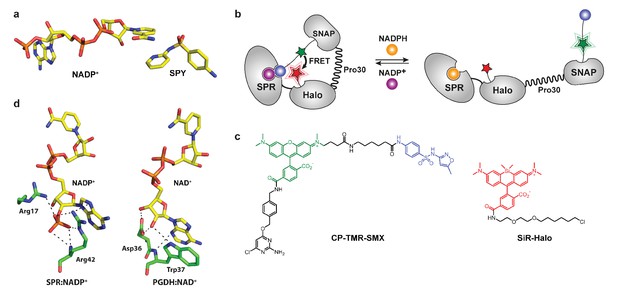
Design of semisynthetic sensors for NADP and NAD+.
(a) Interaction of NADP+ and sulfapyridine in the substrate-binding site of SPR (PDB entry: 4HWK). The pyridine moiety of sulfapyridine (SPY) and the nicotinamide moiety of NADP+ are at a suitable distance (3.3 Å) for efficient π-stacking. (b) The fusion protein SPR-Halo-p30-SNAP is labeled via SNAP-tag with a synthetic molecule containing a FRET donor (green star) and a SPR inhibitor (blue ball, SMX), and via Halo-tag with a FRET acceptor. NADPH (orange ball) and NADP+ (purple ball) compete for the cofactor-binding site of SPR. The sensor can monitor NADPH/NADP+ ratio changes by switching from a closed conformation to an open conformation, with high and low FRET efficiency, respectively. (c) Structures of the synthetic molecules used to constitute the sensor. CP-TMR-SMX contains O4-benzyl-2-chloro-6-aminopyrimidine (CP) for reaction with SNAP-tag, a tetramethylrhodamine (TMR, green) derivative as FRET donor and a tethered sulfamethoxazole (SMX, blue). SiR-Halo is used for the specific labeling of Halo-tag with siliconrhodamine. (d) Interactions of residues contributing to cofactor specificity of the SDR superfamily. NADP(H)-preferring enzymes (e.g. SPR) have two conserved basic residues interacting directly with the 2’-phosphate group of NADP+ (PDB entry: 4HWK). NAD(H)-preferring enzymes (e.g. PGDH) have a conserved aspartic acid interacting in a bidentate manner with the 2’- and 3’-hydroxyl groups of NAD+ (PDB entry: 2GDZ).
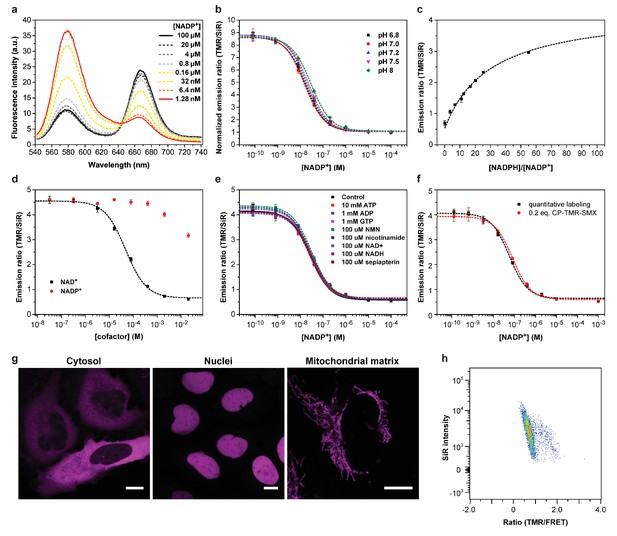
Characterization of NADP- and NAD-Snifit.
(a) Emission spectra of NADP-Snifit titrated with NADP+. TMR and SiR have a maximal emission at 577 and 667 nm, respectively, and the sensor has an isosbestic point at 645 nm. (b) Titrations of NADP-Snifit with NADP+ at various pH ranging from 6.8 to 8.0. The maximum FRET ratio change is 8.9 ± 0.1 fold with a c50 of 29 ± 7 nM. (c) Titration of NADP-Snifit with NADPH/NADP+. The ratio NADPH/NADP+ corresponding to the half maximal sensor response, r50 is 30 ± 3. For the fitting, the upper asymptote is set to the value obtained by adding saturating concentration of sulfamethoxazole (2 mM). (d) Titration of NAD-Snifit labeled with CP-TMR-SMX and SiR-Halo. The maximum ratio change is 7.6 ± 0.2 fold with a c50 of 63 ± 12 µM. (e) Titrations of NADP-Snifit with NADP+ in presence of a fixed concentration of one of the listed different metabolites and structural analogs. (f) Comparative titrations between a quantitatively labeled sensor protein and a sensor protein only labeled with 0.2 equivalent of CP-TMR-SMX. The different fitted parameters from the titration and kinetic experiments are obtained from three independent titrations performed in triplicate. Data represent the mean ± s.d. (g) Confocal images of U2OS cells expressing NADP-Snifit in defined cellular compartments. The images represent the SiR fluorescence of the labeled sensor. Scale bars, 10 µm. (h) Representative gated population of cytosolic NAD-Snifit in U2OS measured by flow cytometry (7000 cells). The graph represents SiR intensity through direct excitation versus FRET ratio.
-
Figure 2—source data 1
- https://doi.org/10.7554/eLife.32638.004
-
Figure 2—source data 2
- https://doi.org/10.7554/eLife.32638.005
-
Figure 2—source data 3
- https://doi.org/10.7554/eLife.32638.006
-
Figure 2—source data 4
- https://doi.org/10.7554/eLife.32638.007
-
Figure 2—source data 5
- https://doi.org/10.7554/eLife.32638.008
-
Figure 2—source data 6
- https://doi.org/10.7554/eLife.32638.009
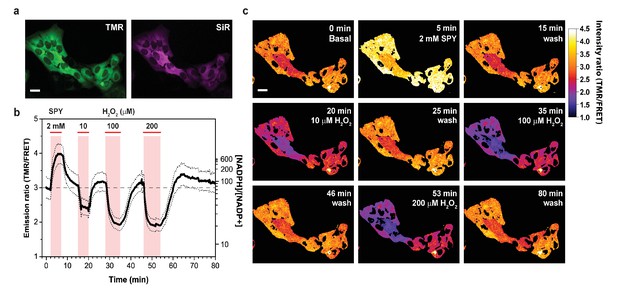
Response of cytosolic NADP-Snifit to H2O2 perfusion.
(a) Pseudocolored widefield images of cytosolic NADP-Snifit expressed and labeled in U2OS cells corresponding to the donor channel (TMR, green) and acceptor channel through direct excitation (SiR, magenta). (b) Time course of the FRET ratio (TMR/FRET) of cytosolic NADP-Snifit upon perfusion of 2 mM sulfapyridine (SPY; to determine the FRET ratio of the sensor in the open state in situ) and increasing concentration of H2O2 (10, 100, 200 µM). The continuous line represents the mean ratio ±s.d. (dotted lines) (n = 10 cells). Free NADPH/NADP+ ratios are represented on the right y-axis. The red bars indicate the time span of perfusion. (c) Ratio images of the cytosolic NADP-Snifit at different time points. Scale bars, 30 µm.
-
Figure 3—source data 1
- https://doi.org/10.7554/eLife.32638.012
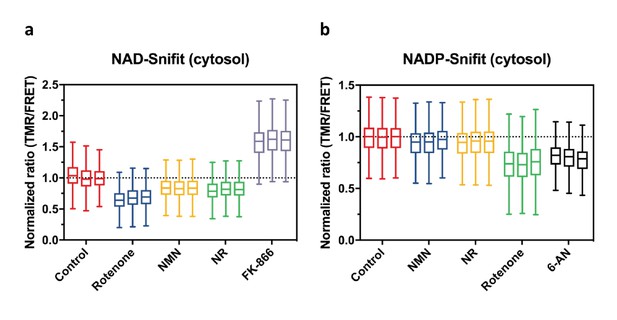
Effects of drugs and NAD biosynthetic precursors on NAD+ and NADPH/NADP+ levels.
FRET ratios (TMR/FRET) as measured by flow cytometry of cytosolic NAD-Snifit (a) and cytosolic NADP-Snifit (b) in U2OS cells after incubation of cells under the conditions specified. For each condition, data from three independent experiments are shown to demonstrate the reproducibility of these measurements. Measured FRET ratios (TMR/FRET) are normalized to untreated control. Abbreviations and conditions: 10 μM Rotenone, 1 mM nicotinamide mononucleotide (NMN), 1 mM nicotinamide riboside (NR), 100 nM FK866, 1 mM 6-aminonicotinamide (6-AN). The Tukey-style box plots represent the 25th and 75th percentiles at the lower and upper box limits and the median as the middle bar. The whiskers extend to ± 1.5 × IQR beyond the limits of the boxes, respectively. The position of the mean is indicated by a solid square. Each data set represent n = 2000–7000 events.
-
Figure 4—source data 1
- https://doi.org/10.7554/eLife.32638.015
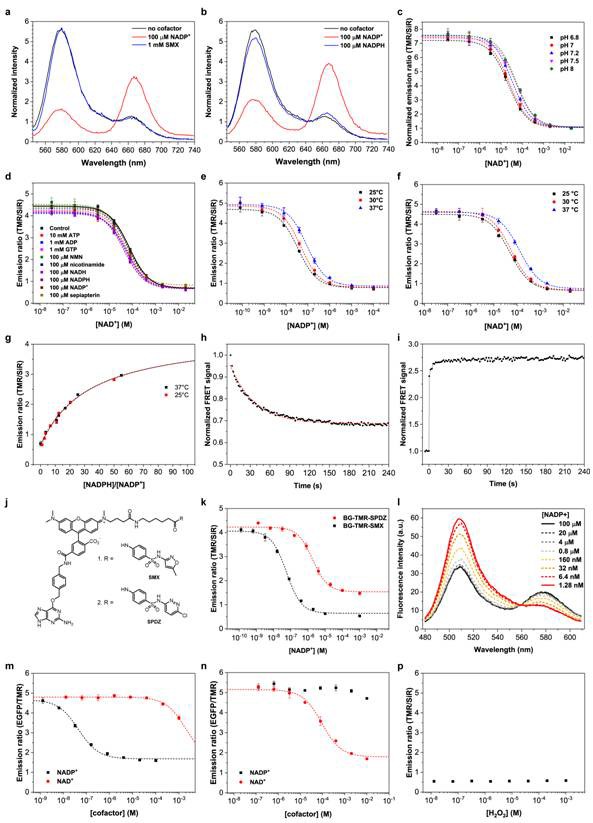
In vitro sensors characterization.
(a) NADP+ binding is a prerequisite for sensor closing. Comparative emission spectra of NADP-Snifit normalized to its isosbestic point (645 nm) in absence of NADP+ (black line), in presence of 100 µM NADP+ (red line) and in presence of 100 µM NADP+ and 1 mM sulfamethoxazole (SMX) (blue line). (b) The intramolecular ligand does not bind to the sensor saturated with NADPH. Emission spectra of NADP-Snifit without NADP+ (black line), after the addition of 1 mM glucose-6-phosphate and 100 µM NADP+ (red line) and finally after a 30 min incubation in presence of 1 nM glucose-6-phosphate dehydrogenase (G6PD). The conversion of NADP+ into NADPH is not fully complete as G6PD is inhibited at high NADPH/NADP+ ratios. However, obtaining pure NADPH is difficult since most commercial stock of NADPH were found to have ~2–3% NADP+ as impurity. (c) Titrations of NAD-Snifit with NAD+ at various pH ranging from 6.8 to 8.0. (d) Titrations of NAD-Snifit with NAD+ in presence of a fixed concentration of one of the listed different metabolites and structurally close molecules and the substrate sepiapterin. (e) Titrations of NADP-Snifit with NADP+ at 25°C, 30°C and 37°C (c50 varies from 35 ± 3 nM to 88 ± 7 nM, from 25°C to 37°C) (f) Titrations of NAD-Snifit with NAD+ at 25°C, 30°C and 37°C (c50 varies from 63 ± 12 µM to 130 ± 14 µM, from 25°C to 37°C). (g) Titrations of NADP-Snifit with varying NADPH/NADP+ ratios at 25°C and 37°C. The r50 of the fitted curves do not change significantly between the two temperatures (r50 is 32 and 33, respectively for 25°C and 37°C). (h) Kinetics of sensor opening. The experiment is conducted by injection of 5 mM NADPH at time zero to the closed sensor saturated with NADP+ (100 nM sensor, 10 µM NADP+). The measured t1/2 fitted with a single-exponential decay is 25 s. (i) Time course of the sensor closing following the injection of 1 mM NADP+ at the zero time point. The experimental set-up does not resolve the closing kinetic for the unsaturated sensor. (j) Chemical structures of BG-TMR-SMX (1) and BG-TMR-SPDZ (2). (k) Titrations of NADP-Snifit labeled either with BG-TMR-SMX or BG-TMR-SPDZ with NADP+. The determined c50 values of the sensor for NADP+ are of 29 ± 7 nM for sulfamethoxazole (SMX) and 1.9 ± 0.3 µM for sulfachloropyridazine (SPDZ) as intramolecular ligand. (l) Emission spectra of the EGFP sensor version SPR(WT)-EGFP-p30-SNAP titrated with NADP+. (m) Titration of SPR-EGFP-p30-SNAP with NADP+ and NAD+. Similarly to NADP-Snifit, the fitted c50 is of 45 nM and ~2 mM (extrapolated), respectively for NADP+ and NAD+. (n) Titration of SPR(D41W42)-EGFP-p30-SNAP with NADP+ and NAD+. The sensor is specific for NAD+ with a fitted c50 of 63 ± 12 µM. (p) NADP-Snifit was titrated up to 1 mM H2O2 with a fixed concentration of NADP+. Unless indicated, the measurements were performed in 50 mM HEPES, 150 mM NaCl, 0.5 mg/mL BSA, pH 7.4 at 25°C. Data represent the mean ± s.d. of titrations performed in triplicate.
-
Appendix 1—figure 1—source data 1
- https://doi.org/10.7554/eLife.32638.019
-
Appendix 1—figure 1—source data 2
- https://doi.org/10.7554/eLife.32638.020
-
Appendix 1—figure 1—source data 3
- https://doi.org/10.7554/eLife.32638.021
-
Appendix 1—figure 1—source data 4
- https://doi.org/10.7554/eLife.32638.022
-
Appendix 1—figure 1—source data 5
- https://doi.org/10.7554/eLife.32638.023
-
Appendix 1—figure 1—source data 6
- https://doi.org/10.7554/eLife.32638.024
-
Appendix 1—figure 1—source data 7
- https://doi.org/10.7554/eLife.32638.025
-
Appendix 1—figure 1—source data 8
- https://doi.org/10.7554/eLife.32638.026
-
Appendix 1—figure 1—source data 9
- https://doi.org/10.7554/eLife.32638.027
-
Appendix 1—figure 1—source data 10
- https://doi.org/10.7554/eLife.32638.028

Live-cell imaging with NAD(P)-Snifit.
Multichannel fluorescence confocal images of NAD(P)-Snifit localized in the cytosol (a), nuclei (b) and mitochondria (c) of U2OS cells. Confocal images of cytosolic NAD(P)-Snifit in HEK293T (d), HeLa (e) and NIH/3T3 (f) cells. (g) Confocal images of NADP-Snifit localized in the mitochondria of NIH/3T3 cells. Widefield images of NAD(P)-Snifit localized in the cytosol (h), nuclei (i) and the mitochondria (j) of U2OS cells and in the cytosol of HEK293T cells (k). All images were taken in full growth medium (DMEM +10% FBS). The different detection channels are represented using pseudocolors: donor channel (TMR, green), FRET channel (red) and the emission of acceptor through direct excitation (SiR, magenta). All images were subject to background correction and the FRET channel was additionally corrected for crosstalk. Scale bars, 20 µm. (l–m) Colocalization of mitochondrial NADP-Snifit with MitoTracker Green. Three color confocal images of MitoTracker Green (MTG, green), mitochondrial localized NADP-Snifit (red) using the acceptor dye channel (SiR) and Hoechst 33342 as nuclear stain (blue) in living U2OS cells. The images were deconvolved using Huygens Essentials package prior to the colocalization analysis. The Pearson’s coefficient between the MTG and SiR channels are 0.80 (l) and 0.90 (m). Scale bars, 10 µm.
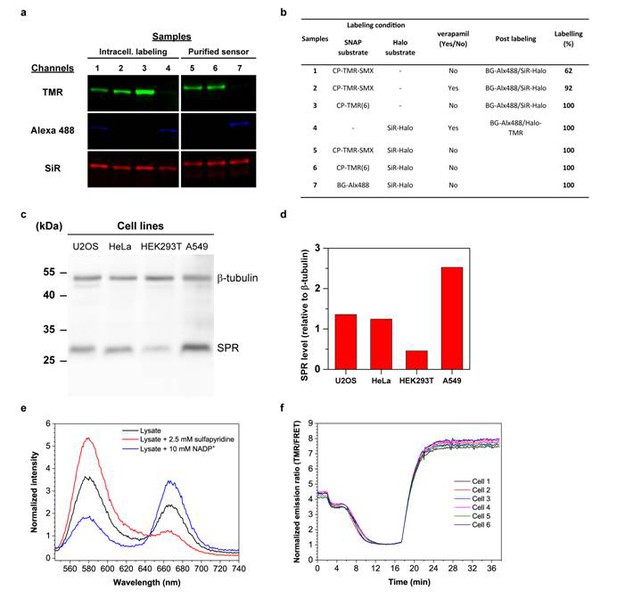
In cellulo sensor characterization.
Intracellular labeling efficiencies. (a) Representative in-gel fluorescence detection of intracellular and in vitro (control) sensor protein labeling. The sensor protein is labeled intracellularly (U2OS cells) with CP-TMR-SMX, CP-TMR(6) or SiR-Halo (samples 1–4) with or without the presence of the efflux pump inhibitor verapamil (10 µM), overnight. The cells were washed and lysed with an excess BG-Alexa(488) and SiR-Halo or Halo-TMR to quantify the unlabeled fraction of SNAP-tag and Halo-tag. As control the purified sensor was labeled in vitro with CP-TMR-SMX/CP-TMR(6)/BG-Alexa(488) and SiR-Halo (samples 5–7). For the quantification of TMR or SiR labeling, the ratio of Alexa(488)/SiR and TMR/SiR of the intracellular samples is calculated relative to the in vitro samples. The results of the labeling efficiency and the description of the samples run on the SDS-PAGE gel can be found in Table b. (c) Comparison of the endogenous SPR level of different cell lines by Western Blot. Western blot of SPR (28 kDa) and β-tubulin (50 kDa) as loading control with different cell lysates revealed by ECL. For each cell lysates, 20 µg total protein were loaded in each well. (d) Representation of the relative expression level of SPR in the different cell lines determined as integrated band intensity normalized to β-tubulin integrated intensity using the displayed blot. (e) The sensor dynamic range is maintained in lysate or in cells. The purified NADP-Snifit is added to a freshly prepared U2OS lysate (0.5 mg/mL protein) to a concentration of 50 nM. The measured TMR/SiR ratio of 1.6 corresponds to a NADPH/NADP+ ratio of 11 in the whole-cell lysate (black line). The sensor was fully open by adding a saturating concentration of free ligand (2.5 mM sulfapyridine) and displays a TMR/SiR ratio of 4.5 (red line). To obtain the fully closed sensor in lysate, 10 mM NADP+ was spiked to the lysate, resulting in a TMR/SiR ratio of 0.5 (blue line). A similar FRET ratio change can be observed for closed sensor in buffer. (f) Semi-stable U2OS cells expressing the nuclear localized NADP-Snifit were used to performed an intracellular sensor calibration. The cells plated on a 12-well plate poly-L-lysine coated coverslip were imaged in HBSS with a widefield microscope. After 2 min, 10 mM NADP+ and 0.001% (w/w) digitonin prepared in HBSS was added to reach the sensor closed state. At 17 min, sulfapyridine was added to a saturating concentration (2 mM) to reach the sensor open state. The dynamic range measured with this widefield microscope was approximately of 8-fold similarly to lysate and buffer measurements.
-
Appendix 1—figure 3—source data 1
- https://doi.org/10.7554/eLife.32638.031
-
Appendix 1—figure 3—source data 2
- https://doi.org/10.7554/eLife.32638.032
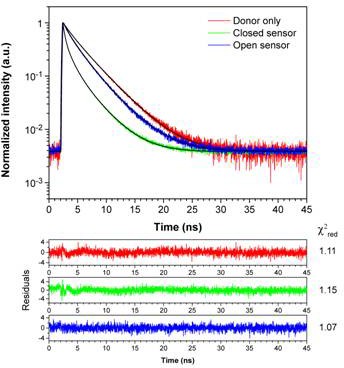
Fluorescence decays of the purified sensor measured by FLIM.
Representative fluorescence decays of NADP-Snifit in buffer (50 mM HEPES, 150 mM NaCl, pH 7.5, 0.5 mg/mL BSA at 37 °C) measured by TCSPC-FLIM. The donor only sample corresponds to purified sensor labeled only with CP-TMR-SMX (red line). The fluorescence decay is fitted with a biexponential model (χ2red = 1.11), yielding an amplitude-weighted average lifetime <τ> of 2.84 ns. The FRET samples are prepared with the addition of 1 mM NADP+ (green line) or 2 mM sulfapyridine (blue line) in the aforementioned buffer to obtain, respectively the closed sensor with highest FRET efficiency (Emax) and the closed sensor with the lowest FRET efficiency (Emin). The fluorescence decays of FRET samples are best fitted with a 3rd-order exponential model. The closed and open sensor conformation yield <τ> of 1.03 (χ2red = 1.15) and 2.4 ns (χ2red = 1.07), respectively. Emax and Emin correspond to 64% and 15% according to the following equation: ; where τDA is the lifetime of the FRET sample (donor + acceptor) and τD is the lifetime of the donor-only sample.
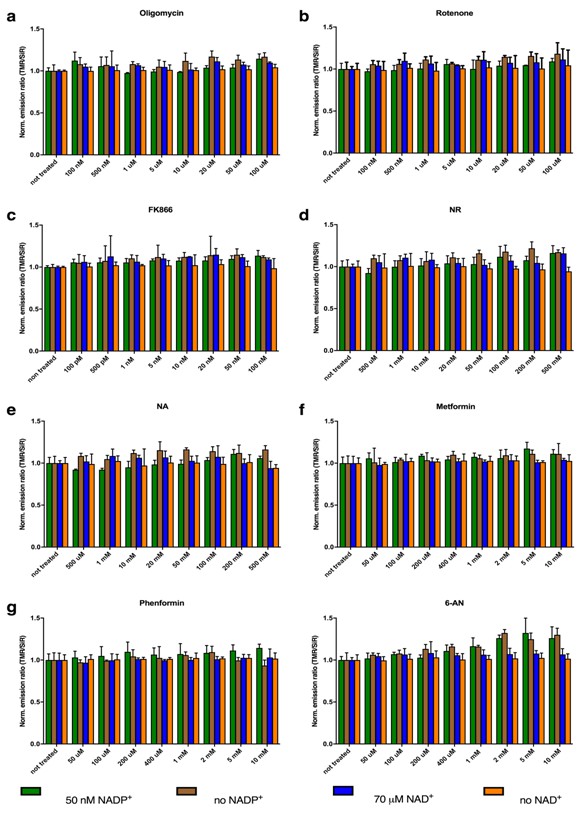
In vitro titration to assess potential interference of the investigated compounds on the sensor’s performance.
NAD(P)-Snifits were incubated with different concentrations of the compounds in the presence of the respective cofactor. Cofactor concentrations were set around the c50 as perturbations on the sensor’s performance should be most prominent in this range. The same experiments were performed in the absence of the cofactor to exclude that the sensors can be closed by the compounds. No significant perturbation of the sensor’s performance was observed. (a) Oligomycin, (b) Rotenone, (c) FK866: (E)-N-[4-(1-benzoylpiperidin-4-yl)butyl]−3-(pyridine-3-yl)acrylamide, (d) NR: nicotinamide riboside, (e) NA: nicotinic acid, (f) Metformin, (g) Phenformin, (h) 6-AN. Three independent experiments were measured and the mean values including ± s.d. are shown.
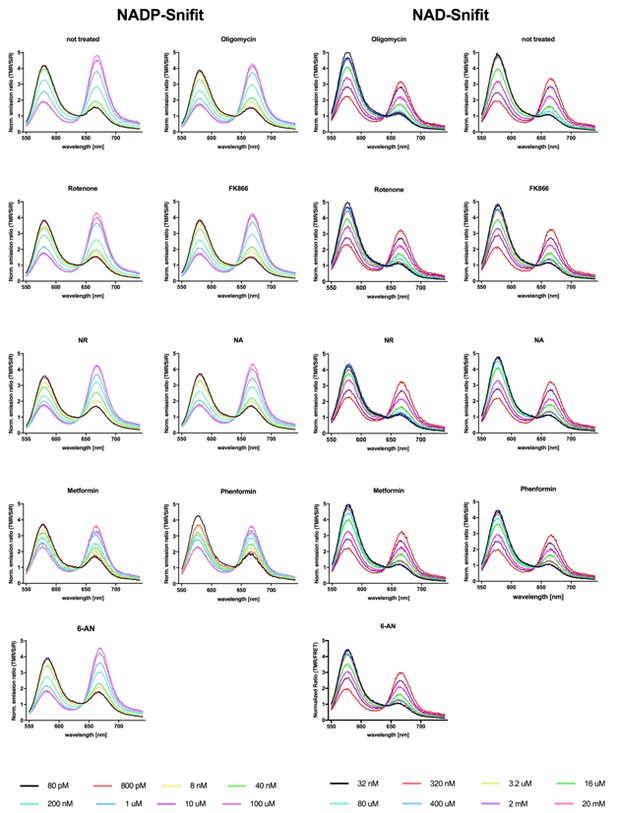
Emission spectra of the NAD(P)-Snifit titrates in the presence of investigated compounds.
The emission spectra were recorded in the presence of the respective compound using the same conditions as for the FACS experiments. No significant alteration of the spectra was observed. Conditions: untreated control, 25 μM Oligomycin, 10 μM Rotenone, 100 nM FK866: (E)-N-[4-(1-benzoylpiperidin-4-yl)butyl]−3-(pyridine-3-yl)acrylamide, 10 mM NR: nicotinamide riboside, 1 mM NA: nicotinic acid, 1 mM Metformin, 1 mM Phenformin.
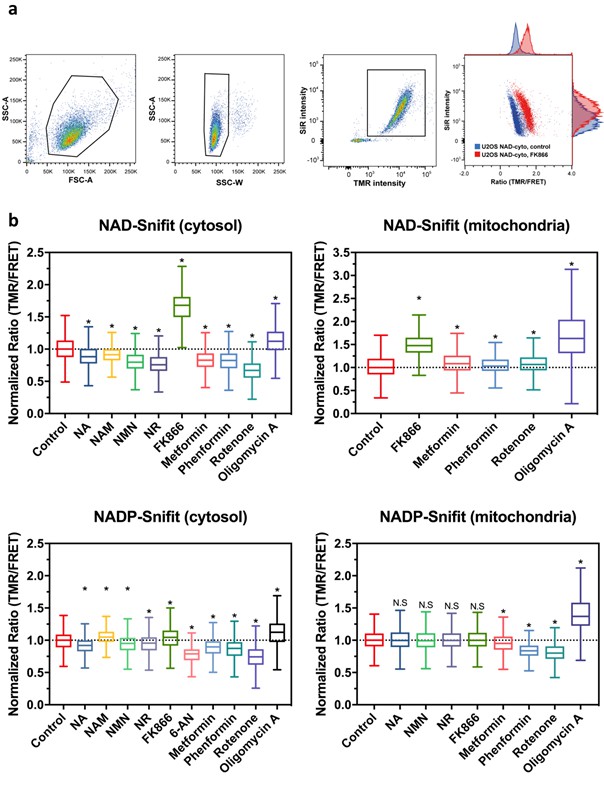
Monitoring NAD+ and NADPH/NADP+by flow cytometry, related to Figure 4 and Table 2.
(A) Representative dot plots of the gating strategy. Live cells and singlets were gated by excluding dead cells and cellular debris (SSC-A vs FCS-A) and doublets or cell clumps (SSC-A vs SSC-W), respectively. Then, only the population of cells with sensor construct fully labeled with CP-TMR-SMX and SiR-Halo were considered for analysis. Example of FRET ratio (TMR/FRET) changes of U2OS cell populations treated for 24 hr with 100 nM of the non-competitive NAMPT inhibitor FK866 (red, significantly increasing the ratio TMR/FRET (i.e. decrease NAD+ level)) compared to the untreated cells (control, blue) (n = 7000 gated cells per condition). (B) Flow cytometry data of cytosolic and mitochondrial NAD(P)-Snifits in U2OS cells after 24 hr incubations under the conditions specified. 1 mM NA, 10 mM NAM, 1 mM NR, 100 nM FK866, 1 mM 6-AN, 1 mM Metformin, 1 mM Phenformin, 10 μM Rotenone and 25 μM Oligomycin A. The results represent the sensor responses measured as FRET ratio (TMR/FRET) normalized to the untreated cell population. The Tukey-style box plots represent the 25th and 75th percentiles at the lower and upper box limits and the median as the middle bar. The whiskers extend to ± 1.5 x IQR beyond the limits of the boxes, respectively. The position of the mean is indicated by a solid square. The data represent one data set for each condition (n = 2000–7000 events). * p<0.05 (Kruskal-Wallis with Dunn’s post-hoc multiple comparison test with respect to control conditions), n.s. = not significant.
-
Appendix 1—figure 7—source data 1
- https://doi.org/10.7554/eLife.32638.037
Tables
Quantification of free NADPH/NADP+ and NAD+ levels in different subcellular compartments of U2OS cells.
https://doi.org/10.7554/eLife.32638.010| NADPH/NADP+ | NAD+ (µM) | |||
|---|---|---|---|---|
| Emission ratio | TCSPC-FLIM | Emission ratio | TCSPC-FLIM | |
| Cytosol | 64.9 ± 26.1 | 55.8 ± 11.7 | 52.8 ± 21.6 | 73.9 ± 7.1 |
| Nucleus | 51.0 ± 16.7 | 40.4 ± 6.7 | n.d. | 117.8 ± 7.2 |
| Mitochondria | 218.7 ± 107.2 | 175.3 ± 57.9 | n.d. | 95.6 ± 7.3 |
-
The values represent the mean ± s.d. of n = 60 and n = 10 cells for the emission ratio and FLIM measurements, respectively. n.d., not determined.
Pharmacological alterations of NAD+ and NADPH/NADP + in U2OS cells measured by flow cytometry.
https://doi.org/10.7554/eLife.32638.013| Normalized FRET ratio (TMR/FRET) | ||||
|---|---|---|---|---|
| Treatment | NAD-Snifit | NADP-Snifit | ||
| Cytosol | Mitochondria | Cytosol | Mitochondria | |
| Control | 1.00 (±0.03) | 1.00 (±0.02) | 1.00 (±0.01) | 1.00 (±0.01) |
| 1 mM NA | 0.91 (±0.01) | n.d. | 0.92 (±0.01) | 1.00 (±0.01)* |
| 10 mM NAM | 0.92 (±0.02) | n.d. | 1.05 (±0.01) | n.d. |
| 1 mM NMN | 0.82 (±0.01) | n.d. | 0.95 (±0.01) | 0.99 (±0.01)* |
| 1 mM NR | 0.80 (±0.02) | n.d. | 0.96 (±0.01) | 1.00 (±0.01)* |
| 100 nM FK866 | 1.61 (±0.06) | 1.48 (±0.04) | 1.05 (±0.01) | 0.99 (±0.01)* |
| 1 mM 6-AN | n.d. | n.d. | 0.80 (±0.02) | n.d. |
| 1 mM Metformin | 0.89 (±0.04) | 1.09 (±0.03) | 0.90 (±0.01) | 0.95 (±0.01) |
| 1 mM Phenformin | 0.79 (±0.05) | 1.13 (±0.06) | 0.88 (±0.01) | 0.83 (±0.01) |
| 10 µM Rotenone | 0.67 (±0.03) | 1.08 (±0.02) | 0.75 (±0.02) | 0.80 (±0.02) |
| 25 µM Oligomycin A | 1.14 (±0.03) | 1.63 (±0.01) | 1.12 (±0.03) | 1.36 (±0.07) |
-
Values represent the average of medians (±s.d.) TMR/FRET ratios of three independent measurements normalized to control condition (n = 3). Control: untreated cells (full growth medium with 25 mM glucose), NA: nicotinic acid, NAM: nicotinamide, NMN: nicotinamide mononucleotide, NR: nicotinamide riboside, FK866: (E)-N-[4-(1-benzoylpiperidin-4-yl)butyl]−3-(pyridin-3-yl)acrylamide, 6-AN: 6-aminonicotinamide.
*The effect of the treatment is not statistically significant compared to the control condition (Kruskal-Wallis with Dunn’s post-hoc multiple comparison test, α = 0.05). n.d., not determined. All compounds were also tested for interactions with the sensor in vitro (Appendix 1—figures 5, 6).
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-SPR (clone EPR9290) | Abcam | Cat#ab157194 |
| Mouse monoclonal anti-β-tubulin (clone 5H1) | BD Biosciences | Cat#556321; RRID: AB_396360 |
| Goat anti-Rabbit secondary antibody, HRP-conjugate | Cell Signaling Technology | Cat#7074; RRID: AB_2099233 |
| Horse anti-Mouse secondary antibody, HRP-conjugate | Cell Signaling Technology | Cat#7076; RRID: AB_330924 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CP-TMR-SMX | This paper | N/A |
| BG-TMR-SMX | This paper | N/A |
| SiR-Halo | This paper | N/A |
| CP-TMR | Johnsson Lab | N/A |
| Sulfapyridine (≥99%) | Sigma-Aldrich | Cat#S6252 |
| Sulfamethoxazole (>98%) | TCI | Cat#S0361 |
| Sulfachloropyridazine | Sigma-Aldrich | Cat#S9882 |
| (±)-Verapamil hydrochloride (≥99%) | Sigma-Aldrich | Cat#V4629 |
| H2O2 (30% (w/w), puriss. p.a.) | Sigma-Aldrich | Cat#31642 |
| 2-Deoxy-D-glucose (≥99%) | Sigma-Aldrich | Cat#D6134 |
| 6-aminonicotinamide (99%) | Sigma-Aldrich | Cat#A68203 |
| Resveratrol (>99%) | TCI | Cat#R0071 |
| Nicotinic acid (≥99.5%) | Sigma-Aldrich | Cat#72309 |
| Nicotinamide (>98%) | Sigma-Aldrich | Cat#N0636 |
| β-Nicotinamide mononucleotide (95–100%) | Sigma-Aldrich | Cat#N3501 |
| Nicotinamide riboside | Auwerx Lab, EPFL | N/A |
| FK866 hydrochloride hydrate (≥98%) | Sigma-Aldrich | Cat#F8557 |
| Metformin (97%) | Sigma-Aldrich | Cat#D150959 |
| Phenformin | Sigma-Aldrich | Cat#P7045 |
| Rotenone (≥95%) | Sigma-Aldrich | Cat#R8875 |
| Oligomycin A (≥95%) | Sigma-Aldrich | Cat#75351 |
| NADPH tetrasodium salt (≥97%) | Roche | Cat#10621692001 |
| NADP+ disodium salt (≥97%) | Roche | Cat#10128058001 |
| NADH disodium salt (≥95%) | AppliChem | Cat#A1393,0001 |
| NAD+ free acid (100%) | Roche | Cat#10127965001 |
| ATP disodium salt (≥98%) | AppliChem | Cat#A1348,0005 |
| ADP sodium salt (≥95%) | Sigma-Aldrich | Cat#A2754 |
| GTP sodium salt hydrate (≥95%) | Sigma-Aldrich | Cat#G8877 |
| L-sepiapterin | Cayman | Cat#81650 |
| MitoTracker Green FM | Life Technologies | Cat#M7514 |
| Hoechst 33342 | Life Technologies | Cat#H1399 |
| Propidium iodide (≥94%) | Sigma-Aldrich | Cat#81845 |
| Experimental Models: Cell Lines | ||
| U-2 OS (Human osteosarcoma) | ECACC | Cat#92022711 |
| HEK-293T (Human embryonic kidney) | ATCC | Cat#CRL-3216 |
| NIH/3T3 (Mouse embroynic fibroblast) | ATCC | Cat#CRL-1658 |
| HeLa (Human cervix epitheloid carcinoma) | ATCC | Cat#CCL-2 |
| A549 (Human lung carcinoma) | ECACC | Cat#86012804 |
| Recombinant DNA | ||
| pET-51b(+) | Novagen | 71553 |
| pEBTet | (Bach et al., 2007) | N/A |
| pET-51b(+)_NADP | This paper | N/A |
| pET-51b(+)_NAD | This paper | N/A |
| pEBTet_NADP-cyto | This paper | N/A |
| pEBTet_NADP-nucl | This paper | N/A |
| pEBTet_NADP-mito | This paper | N/A |
| pEBTet_NAD-cyto | This paper | N/A |
| pEBTet_NAD-nucl | This paper | N/A |
| pEBTet_NAD-mito | This paper | N/A |
| Software and Algorithms | ||
| OriginPro 9 | OriginLab Corporation | http://www.originlab.com/ |
| PyMOL | Schrödinger, LLC | https://www.pymol.org/ |
| FIJI (ImageJ) | (Schindelin et al., 2012) | https://fiji.sc/ |
| SymPhoTime 64 | PicoQuant | https://www.picoquant.com/ |
| Huygens Essential | Scientific Volume Imaging | https://svi.nl/HuygensEssential |
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/ |
| R 3.4.0 | R Core Team, 2017 | https://www.r-project.org/ |
| Other | ||
| Leica TCS SP8 X confocal microscope - PicoHarp 300 (PicoQuant) TCSPC module | Leica/PicoQuant | http://www.leica-microsystems.com https://www.picoquant.com/ |
| IN Cell Analyzer 2200 automated widefield microscope | GE Healthcare Life Sciences | http://www.gelifesciences.com/ |
| Leica DMI6000B widefield microscope | Leica | http://www.leica-microsystems.com |
Properties of TMR and SiR substrates.
https://doi.org/10.7554/eLife.32638.038| Substrate name | Reaction rate constant (M−1 s−1) (± SD) | Excitation maximum (nm) | Emission maximum (nm) | Lifetime* (ns) |
|---|---|---|---|---|
| BG-TMR(6) | 114’706 (± 5082) | 555 | 577 | 2.2 |
| CP-TMR(6) | 109’278 (± 3338) | 555 | 577 | 2.2 |
| BG-TMR-SMX | 25’836 (± 2307) | 555 | 577 | 2.7 |
| CP-TMR-SMX | 38’847 (± 2307) | 555 | 577 | 2.7 |
| SiR-Halo | >250’000 10 | 650 | 667 | 3.1 |
-
Values for the excitation/emission maxima and the lifetimes were measured with the protein-bound fluorophore.
*The lifetimes correspond to the amplitude-weighted average lifetime measured at 22 °C in HEPES buffer.
Intracellular NAD(P)-Snifit concentration.
https://doi.org/10.7554/eLife.32638.039| Localization | Concentration (µM) (± SD) | N cells |
|---|---|---|
| Cytosol | 1.6 (± 1.4) | 84 |
| Nucleus | 4.0 (± 3.3) | 51 |
| Mitochondria | 4.7 (± 1.3) | 49 |
-
Concentrations were determined in U2OS cells using a confocal fluorescence microscope by singly labeled the sensor construct with SiR-Halo and CP-TMR-SMX and comparing with the purified sensor calibration curves in buffer using identical microscope settings.
Quantification of the cytosolic free [NADPH]/[NADP+] and [NAD+] in different cell lines by TCSPC-FLIM.
https://doi.org/10.7554/eLife.32638.040| Cell lines | [NADPH]/[NADP+] | [NAD+] (µM) |
|---|---|---|
| U2OS | 55.8 ± 11.7 | 73.9 ± 7.1 |
| HEK293T | 21.6 ± 3.4 | 63.6 ± 4.5 |
| NIH/3T3§ | 39.5 ± 12.4 | 44.6 ± 11.2 |
| HeLa | 75.0 ± 11.8 | 49.8 ± 2.4 |
Estimated free [NAD+] and [NADPH]/[NADP+] of pharmacologically treated U2OS cells measured by flow cytometry.
https://doi.org/10.7554/eLife.32638.041| Free [NAD+] (µM) | Free [NADPH]/[NADP+] ratio | |||
|---|---|---|---|---|
| Compound | Cytosol | Mitochondria | Cytosol | Mitochondria |
| Control | 132 (± 29) | 96 (± 20) | 72 (± 8) | 120 (± 14) |
| 1 mM NA | 162 (± 33) | n.d. | 52 (± 6) | 119 (± 14)† |
| 10 mM NAM | 146 (± 30) | n.d. | 87 (± 10) | n.d. |
| 1 mM NMN | 198 (± 41) | n.d. | 59 (± 7) | 114 (± 14)† |
| 10 mM NR | 210 (± 43) | n.d. | 60 (± 7) | 120 (± 14)† |
| 100 nM FK866 | 42 (± 9) | 34 (± 7) | 88 (± 10) | 116 (± 14)† |
| 1 mM 6-AN | n.d. | n.d. | 35 (± 5) | n.d. |
| 1 mM Metformin | 168 (± 35) | 80 (± 17) | 49 (± 6) | 90 (± 10) |
| 1 mM Phenformin | 213 (± 45) | 72 (± 15) | 45 (± 6) | 54 (± 6) |
| 10 µM Rotenone | 300 (± 64) | 81 (± 17) | 29 (± 4) | 47 (± 6) |
| 25 µM Oligomycin A | 101 (± 21) | 24 (±5) | 121 (± 24) | open sensor* |
-
Values represent the mean estimated concentrations and ratios (± SD) of three independent measurements performed in triplicate. The TMR/FRET ratios were converted into concentration using Equations 7 and 8, where Rmax was determined in situ by incubating 10 min the cells with 2 mM sulfapyridine. Rmin was calculated from the in vitro maximum FRET ratio change ΔRmax (Rmin = Rmax/ΔRmax). c50 and r50 were determined from in vitro titrations at 25 °C. Control: untreated cells (full growth medium with 25 mM glucose), NA: nicotinic acid, Nam: nicotinamide, NMN: nicotinamide mononucleotide, NR: nicotinamide riboside, FK866: (E)-N-[4-(1-benzoylpiperidin-4-yl)butyl]−3-(pyridin-3-yl)acrylamide, 6-AN: 6-aminonicotinamide.
*The sensor reached full opening with this treatment [NADPH]/[NADP+] ≥ 300.
-
†The effect of the treatment is not statistically different compared to the control condition (p ≥ 0.05 using a two-tailed Student’s t-test). n.d., not determined.
In vitro lifetime characterization of NADP-Snifit.
https://doi.org/10.7554/eLife.32638.042| Sample | <τ> (ns) ± SD | E (%) ± SD |
|---|---|---|
| Donor-only | 2.84 ± 0.01 | - |
| FRET (closed sensor) | 1.03 ± 0.01 | 63.9 ± 0.1 |
| FRET (open sensor) | 2.40 ± 0.01 | 15.3 ± 0.1 |
-
The ‘donor only’ sample represents the purified sensor singly labeled with CP-TMR-SMX. The FRET samples corresponding to the closed and open sensor state were prepared respectively with 1 mM NADP+ and 2 mM sulfapyridine. The amplitude-weighted average lifetimes <τ> are represented as mean ± SD of triplicates. All samples were measured in buffer (50 mM HEPES, 150 mM NaCl, 0.5 mg/mL BSA, pH 7.5) at 37 °C. From the obtained lifetimes, the FRET efficiency (E) of the closed and open sensor was calculated.
Determination of FRET efficiency in U2OS cells.
https://doi.org/10.7554/eLife.32638.043| Sensors | Localization | <τ> (ns) ± SD | E (%) ± SD (Basal) | Emin (%) ± SD (2 mM SPY) | ||
|---|---|---|---|---|---|---|
| Donor only | FRET, basal | FRET, 2 mM SPY | ||||
| NADP-Snifit | Cytosol | 2.80 ± 0.02 | 2.12 ± 0.06 | 2.35 ± 0.04 | 24.2 ± 0.7 | 16.0 ± 0.3 |
| Nucleus | 2.69 ± 0.03 | 1.98 ± 0.04 | 2.28 ± 0.04 | 26.3 ± 0.6 | 15.5 ± 0.4 | |
| Mitochondria | 2.55 ± 0.05 | 2.08 ± 0.03 | 2.16 ± 0.02 | 18.2 ± 0.4 | 15.2 ± 0.3 | |
| NAD-Snifit | Cytosol | 2.89 ± 0.04 | 2.19 ± 0.02 | 2.42 ± 0.02 | 24.3 ± 0.4 | 16.3 ± 0.3 |
| Nucleus | 2.66 ± 0.03 | 2.11 ± 0.04 | 2.49 ± 0.06 | 20.6 ± 0.4 | 6.5 ± 0.2 | |
| Mitochondria | 2.63 ± 0.03 | 2.02 ± 0.02 | 2.30 ± 0.06 | 23.0 ± 0.3 | 12.2 ± 0.3 | |
-
The data represent the amplitude-weighted average lifetime <τ> as mean ± SD (N = 10) measured in living U2OS cells in full growth medium (DMEM +10% FBS) at 37 °C. The ‘donor-only’ sample was obtained by single labeling of the sensor constructs with CP-TMR-SMX. The FRET samples are labeled with both CP-TMR-SMX and SiR-Halo. The cells labeled with both fluorophores were first measured without treatment to obtain their basal fluorescence lifetime, then the same cells were measured again after the treatment with 2 mM sulfapyridine (SPY) to obtain the fully sensor open state. The correlated FRET efficiencies (E) were calculated for each conditions.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32638.016
-
Appendix 1—figure 1—source data 1
- https://doi.org/10.7554/eLife.32638.019
-
Appendix 1—figure 1—source data 2
- https://doi.org/10.7554/eLife.32638.020
-
Appendix 1—figure 1—source data 3
- https://doi.org/10.7554/eLife.32638.021
-
Appendix 1—figure 1—source data 4
- https://doi.org/10.7554/eLife.32638.022
-
Appendix 1—figure 1—source data 5
- https://doi.org/10.7554/eLife.32638.023
-
Appendix 1—figure 1—source data 6
- https://doi.org/10.7554/eLife.32638.024
-
Appendix 1—figure 1—source data 7
- https://doi.org/10.7554/eLife.32638.025
-
Appendix 1—figure 1—source data 8
- https://doi.org/10.7554/eLife.32638.026
-
Appendix 1—figure 1—source data 9
- https://doi.org/10.7554/eLife.32638.027
-
Appendix 1—figure 1—source data 10
- https://doi.org/10.7554/eLife.32638.028
-
Appendix 1—figure 3—source data 1
- https://doi.org/10.7554/eLife.32638.031
-
Appendix 1—figure 3—source data 2
- https://doi.org/10.7554/eLife.32638.032
-
Appendix 1—figure 7—source data 1
- https://doi.org/10.7554/eLife.32638.037





