CK1/Doubletime activity delays transcription activation in the circadian clock
Figures
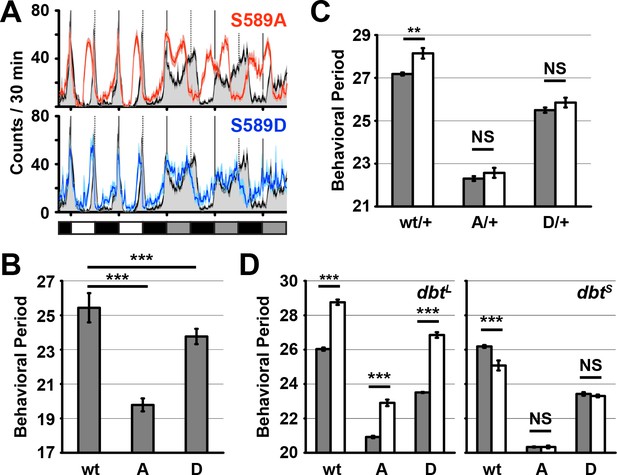
Residue substitutions of S589 differently affect behavioral rhythmicity.
(A) Transgenic wild-type per (black line), S589A per (red line) or S589D per (blue line) were expressed in a per0 genetic background and monitored for behavior in a 12:12 light dark cycle followed by constant darkness and activity recorded and plotted. The solid vertical lines demark days, while the dotted vertical lines represent the light-to-dark transition point. White, black and grey boxes represent lights on, lights off and subjective day during lights off, respectively. N = 27–28. Each data point is the mean ± SEM. Thickness of the curve reflects SEM. (B) Flies described in panel A quantified for behavioral periodicity in constant darkness. wt: wild type, A: S589A, D: S589D. N = 25–27. Bars represent means ± SD. ***p<0.005. Significance was measured by an unpaired, two-tailed t-test. See Supplementary file 1 for values. (C) The behavioral period of heterozygous flies carrying the transgenic per variant (as indicated) in the absence of DBT overexpression (grey bars, per0; attP40{per} / +; UAS-dbt) or with DBT overexpression in clock neurons (white bars, per0; attP40{per}/tim-UAS-Gal4; UAS-dbt) were quantified and plotted. N = 19. Bars represent means ± SD. **p<0.01. NS, not statistically significant. Significance was measured by t-test. See Supplementary file 1 for values. (D) The behavioral period of per transgenic flies in a dbtL or dbtS genetic background (white bars) or wild type dbt background (grey bars) were quantified and plotted. N = 8–24. Bars represent means ± SD. ***p<0.005. NS, not statistically significant. Significance was measured by t-test. See Supplementary file 1 for values.
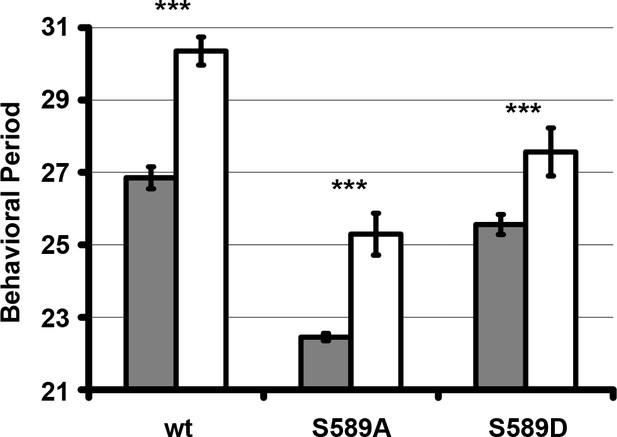
Residue substitutions of S589 do not genetically interact with O-Glc-NAc Transferase (OGT).
The behavioral period of heterozygous flies carrying the transgenic per variant (as indicated) in the absence of DBT overexpression (grey bars, per0; [per] / +; +) or with DBT overexpression in clock neurons (white bars, per0; [per]/tim-UAS-Gal4, UAS-OGT; +) were quantified and plotted. N = 7–16. Bars represent means ±SD. ***p<0.001. Significance was measured by t-test. See Supplementary file 1 for values.
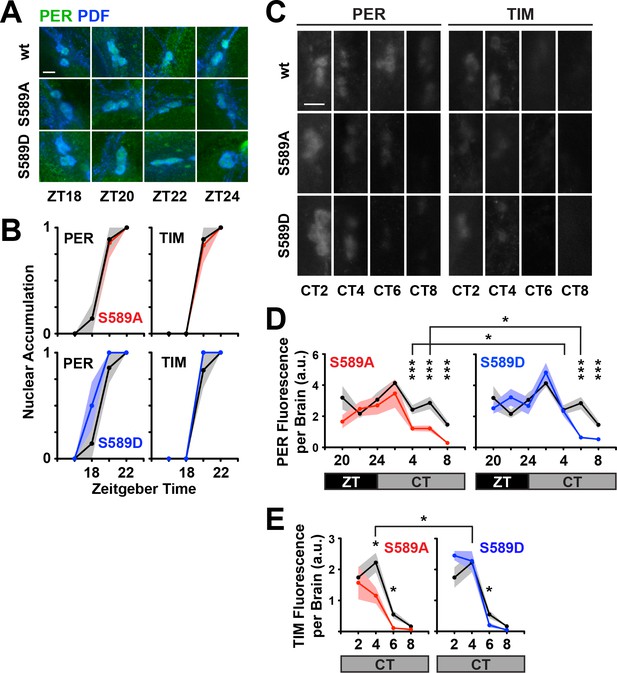
Residue substitutions of S589 do not affect nuclear entry, advance PER degradation in small ventral lateral neurons in vivo.
(A) Fly brains were collected at the indicated zeitgeber time points (ZT) and fixed. Representative images of s-LNvs stained for PER (green) and PDF (blue) are shown. The scale bar represents 5 μm. (B) Stained fly brains were visually scored to determine PER/TIM nuclear accumulation. PER or TIM staining that is predominantly nuclear or cytoplasmic in the small LNvs were valued 1 or 0, respectively. Six to ten fly brains were averaged for each mutant (S589A, red line; S589D, blue line) at the indicated time points, and compared to wild type control (black line). Each data point represents the mean, ±SEM. Thickness of the curve reflects SEM. (C) Fly heads were collected at the indicated circadian time points (CT; constant darkness). Representative images of s-LNvs stained for PER and TIM are shown. PDF stain is omitted for clarity of PER/TIM signal intensity. The scale bar represents 5 μm. (D) Stained fly brains were quantified for PER expression in small LNvs. Fluorescence signal from 6 to 10 fly brains were imaged and quantified (arbitrary units, a.u.) for each time point, normalized to an internal standard and the two hemispheres averaged to represent the brain. Mutant PER expression (S589A, red line; S589D, blue line) and wild type PER (black line) was plotted and compared. Times are measured in ZT (black box) or in CT, subjective day (grey box). Each data point represents the mean, ±SEM. Thickness of the curve reflects SEM. Black line compares the S589A and S589D mutants. *p<0.05. ***p<0.005. Data points with no asterisks are not statistically significant. Significance was measured by t-test. (E) Stained fly brains were quantified for TIM fluorescence signal as described in panel D. Each data point represents the mean, ±SEM, plotted across CT. Thickness of the curve reflects SEM. Black line compares the S589A and S589D mutants. *p<0.05. Significance was measured by t-test.
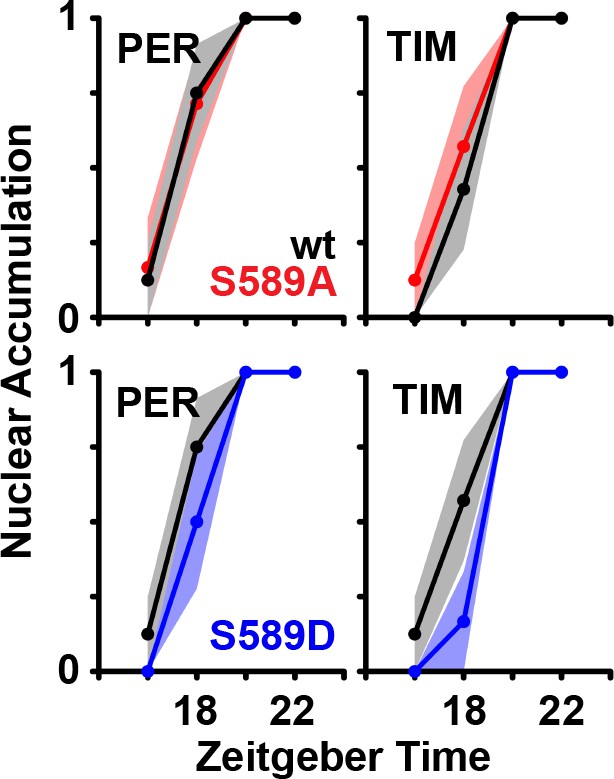
Residue substitutions of S589 do not affect nuclear accumulation in the large ventral lateral neurons in vivo.
Stained fly brains were visually scored to determine PER/TIM nuclear accumulation. PER or TIM staining that is predominantly nuclear or cytoplasmic in the small LNvs were valued 1 or 0, respectively. Six to ten fly brains were averaged for each mutant (S589A, red line; S589D, blue line) at the indicated time points, and compared to wild type (black line). Times are measured in ZT. Each data point represents the mean, ±SEM. Thickness of the curve reflects SEM.
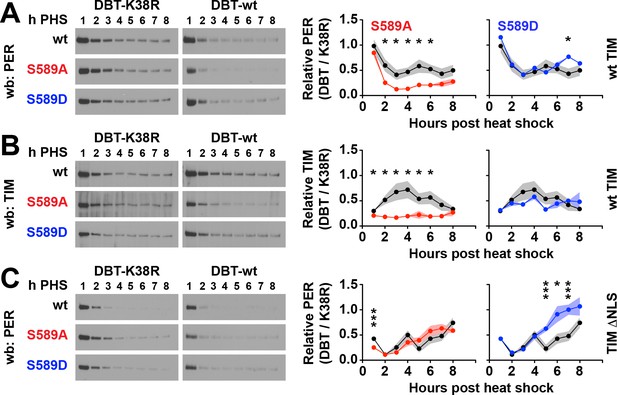
Residue substitutions of S589 determine PER/TIM stability in cultured cells.
S2 cells expressing per driven by a heat-shock promoter, and dbt and tim driven by actin promoter were collected each hour post heat shock (h PHS) in a pulse-chase experiment using cycloheximide. (A) Wild type, S589A or S589D per variants were co-expressed in S2 cells with wild type (wt) TIM and either wild type DBT (DBT-wt) or mutant DBT lacking ATPase activity (DBT-K38R). Representative western blots (wb) are shown. PER signal was quantified and normalized to either DBT-wt or DBT-K38R signal, and the ratios plotted. S589A (red) and S589D (blue) was compared to wild type PER (black). N = 4. Each data point represents the mean, ±SEM. Thickness of the curve reflects SEM. *p<0.05. ***p<0.005. Data points without asterisks are not statistically significant. Significance was measured by t-test. (B) The same analysis conducted in panel A was repeated for TIM co-expressed with PER, S589A or S589D, and DBT-wt or DBT-K38R. (C) The same analysis conducted in panel A was repeated for PER, with a mutant form of TIM lacking a nuclear localization signal (TIM ΔNLS).
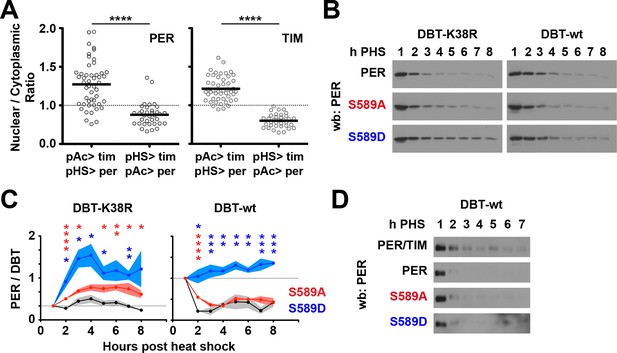
PER/TIM nuclear accumulation in S2 cells is promoter-dependent, PER stability is plotted as a PER/DBT ratio.
(A) S2 cells were transfected with plasmids that express per-mCherry (PER) or tim-YFP (TIM) fusion genes by actin promoter (pAc>) or heat shock promoter (pHS>) as indicated. Cells were imaged 2 hr post heat shock and fluorescence signal in the nucleus and cytoplasm quantified, and the nuclear/cytoplasmic ratios plotted as shown. The black bar represents the mean. The dotted line represents the value at which nuclear and cytoplasmic signals are equal. ****p<0.001. Significance was measured by t-test. (B) Representative blot of mutant DBT (DBT-K38R) or wild type DBT (DBT-wt) co-expressed with TIM and the indicated PER variant, as described in Figure 3. (C) Quantified PER levels are quantified and plotted as a PER/DBT ratio. Left and right panels represent PER co-expressed with DBT-K38R and DBT-wt, respectively. Mutant PER (S589A, red line; S589D, blue line) are compared to wild type PER (black line). N = 4. Each data point represents the mean, ±SEM. Thickness of the curve reflects SEM. Asterisks represent significance of difference between the indicated mutant (S589A, red; S589D, blue) and wild type. *p<0.05. **p<0.01. ***p<0.005. ****p<0.001. Significance was measured by t-test. (D) Representative blots of PER expression in S2 cells with wild type DBT and no TIM. Blots were overexposed to ensure observable protein signal in the first hour.
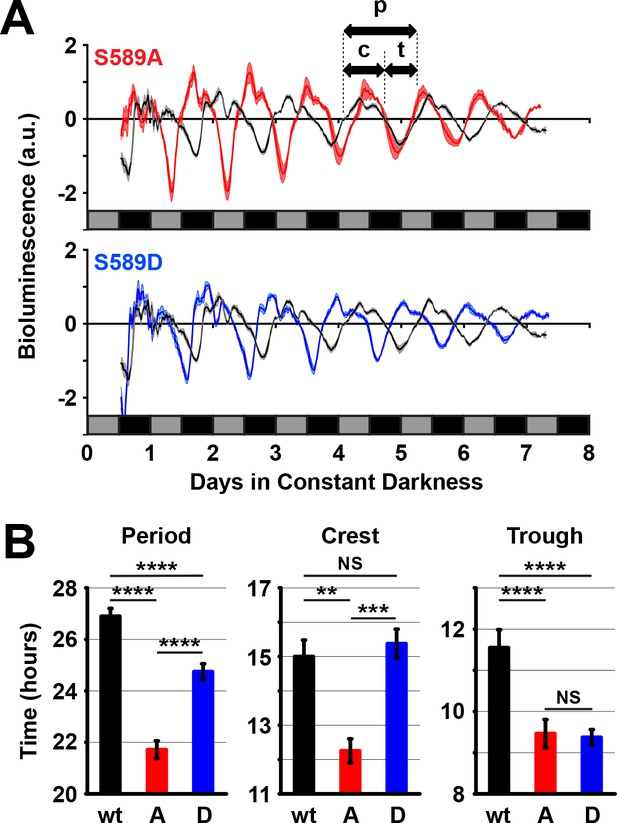
589 substitutions differently regulate per promoter activity in vivo.
(A) Flies expressing the indicated transgenic per variant in a per0 background and the luciferase gene driven by the per promoter were measured for bioluminescence activity in constant darkness. Grey boxes represent subjective day, black boxes represent subjective night. S589A (red line) and S589D (blue line) were compared to wt per rescue (black line) flies. N = 5 groups of 80 flies. Thickness of the curve reflects SEM. Double-headed arrows use the wild type signal to illustrate a full period of bioluminescence (p), the width of the crest (c) or the width of the trough (t). (B) The full period of bioluminescence, and the crest and trough widths measured in panel A are plotted. w: wild type PER (black bars), A: S589A (red bars), D: S589D (blue bars). Bars represent means, ±SEM. **p<0.01. ***p<0.005. ****p<0.001. NS: not statistically significant. Significance was measured by t-test.
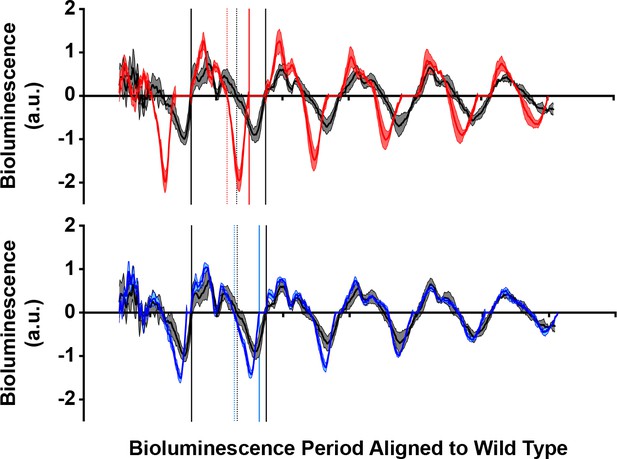
Realignment of bioluminescence period.
Bioluminescence data in Figure 4A replotted to align the start of each crest on each day to emphasize the differences in crest and trough width each day. Solid lines depict the bounds of a full period. Dotted lines bisect the curve at the point of intersecting the x-axis, separating crest and trough. Lines and curves are color coded to represent wild type PER (black), S589A (red) and S589D (blue).
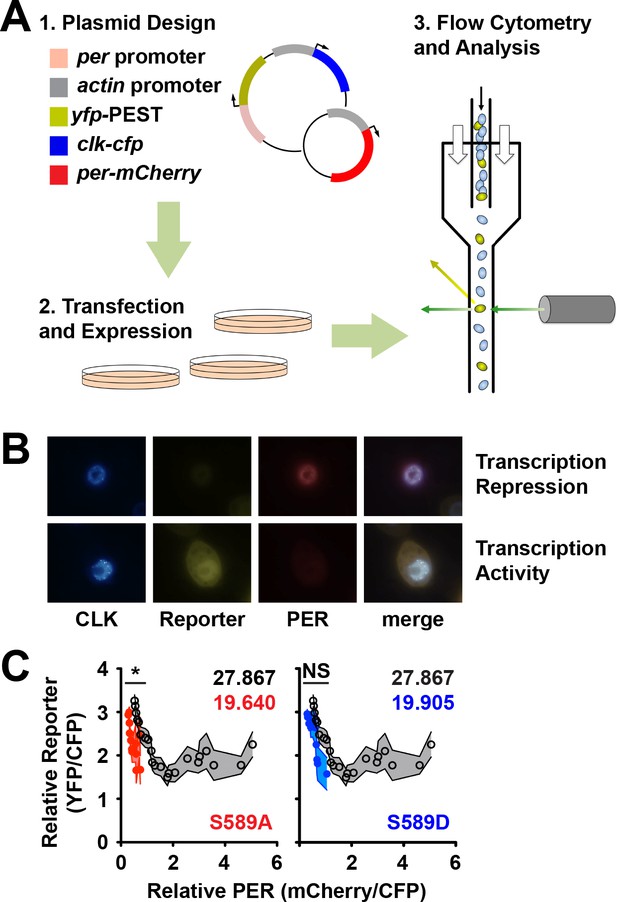
Fluorescence based transcription assay allows simultaneous measurement of transcription inhibition and protein stability.
(A) Flow chart of the principles behind the fluorescence-based transcription assay. (1) Actin promoter (grey) driven clk-cfp (blue) and per promoter (light red) driven yfp (yellow) is cloned into the same expression plasmid and co-expressed in S2 cells with plasmid carrying actin promoter (grey) driven per-mCherry (red). (2) Cells are transfected and then (3) analyzed for fluorescence signal by flow cytometry. (B) Representative images of S2 cells expressing plasmids described in panel A that express high PER and low reporter (top row) and low PER and high reporter (bottom row). Expression of dCLK (blue), reporter (yellow), PER (red) is shown, with a merged image of all three signals. (C) CFP (dCLK)-positive cells were quantified for YFP (reporter) and mCherry (PER) expression using flow cytometry. mCherry signal was binned, and both mCherry and YFP signals were normalized to CFP signal and plotted as shown. S589A (red dots) and S589D (blue dots) were compared to wild-type PER (open black circles). Each data point represents the mean of the binned cells, ±SD. Thickness of the curve reflects SD. *p<0.05. NS, not statistically significant. Significance was measured by Mann-Whitney test. Colored inset numbers represent corresponding PER fluorescence signal per cell in arbitrary units as a measure of PER stability (Black numbers represent wild type PER stability).
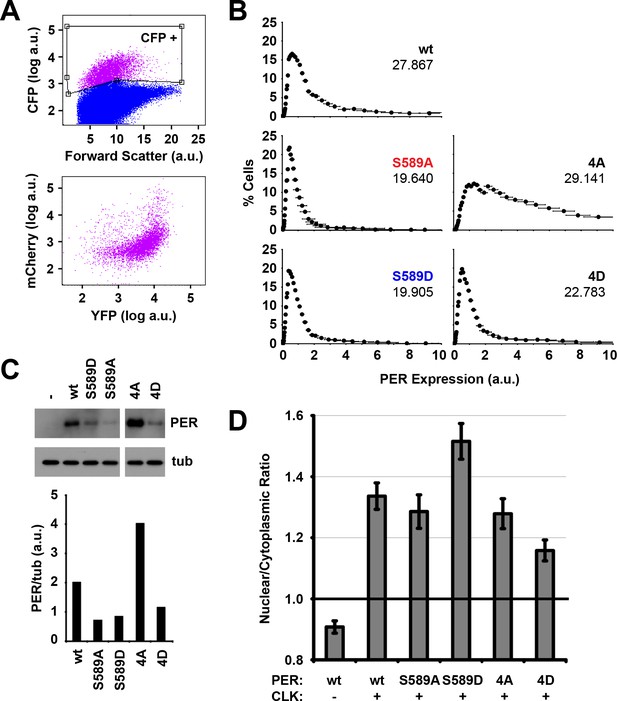
Protein stability and nuclear accumulation in the transcription inhibition assay.
(A) Scatter plot of S2 cells that were gated for CFP expression (CFP+), and re-plotted to demonstrate YFP and mCherry expression. (B) Cells analyzed in Figure 4 were plotted for mCherry fluorescence (PER signal) as a function of cell percentage. Inset: mean values of PER/cell, calculated by averaging total PER signal and dividing by cell number. (C) Western blots probed for mCherry (PER) and tubulin control (tub) were plotted and quantified. PER variants are wt: wild type PER, S589A, S589D, 4A: PER 4A, 4D: PER 4D. (D) mCherry signal from S2 cells expressing wild type or mutant PER, in the presence (+) or absence (-) of dCLK are plotted as a Nuclear/Cytoplasmic ratio of PER fluorescence signal. N = 40–50 cells. Each bar represents the mean, ±SEM.
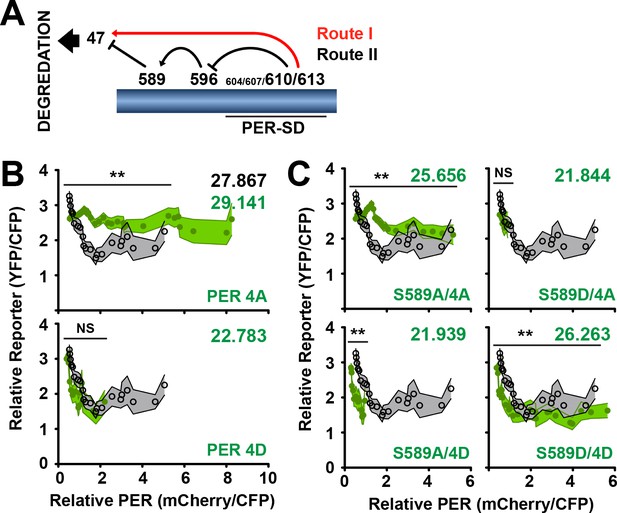
S589 and the PER Short Downstream Domain (PER-SD) cooperate to regulate transcriptional inhibition and PER protein stability.
(A) The phosphorylation cascade model of PER. Phosphorylation of the PER-SD region blocks S589 phosphorylation through an intermediary site (S596) while promoting S47 phosphorylation. S589 phosphorylation blocks S47 phosphorylation. S47 phosphorylation promotes PER degradation. Routes I and II are labeled as they are referred to in the text. (B) The transcription analysis described in Figure 5 is applied to per mutants PER 4A (S604A/S607A/T610A/S613A) and PER 4D (S604D/S607D/T610D/S613D) (green circles) and compared to wild type (open black circles). Each data point represents the mean of the binned cells, ±SD. Thickness of the curve reflects SD. **p<0.01. NS, not statistically significant. Significance was measured by Mann-Whitney test. Colored inset numbers represent corresponding PER fluorescence signal per cell in arbitrary units as a measure of PER stability (Black numbers represent wild type PER stability). (C) The transcription analysis is applied to per mutants S589A/4A, S589A/4D, S589D/4A, S589D/4D (green circles) and compared to wild type (open black circles). Each data point represents the mean of the binned cells, ±SD. Thickness of the curve reflects SD. **p<0.01. NS, not statistically significant. Significance was measured by Mann-Whitney test.
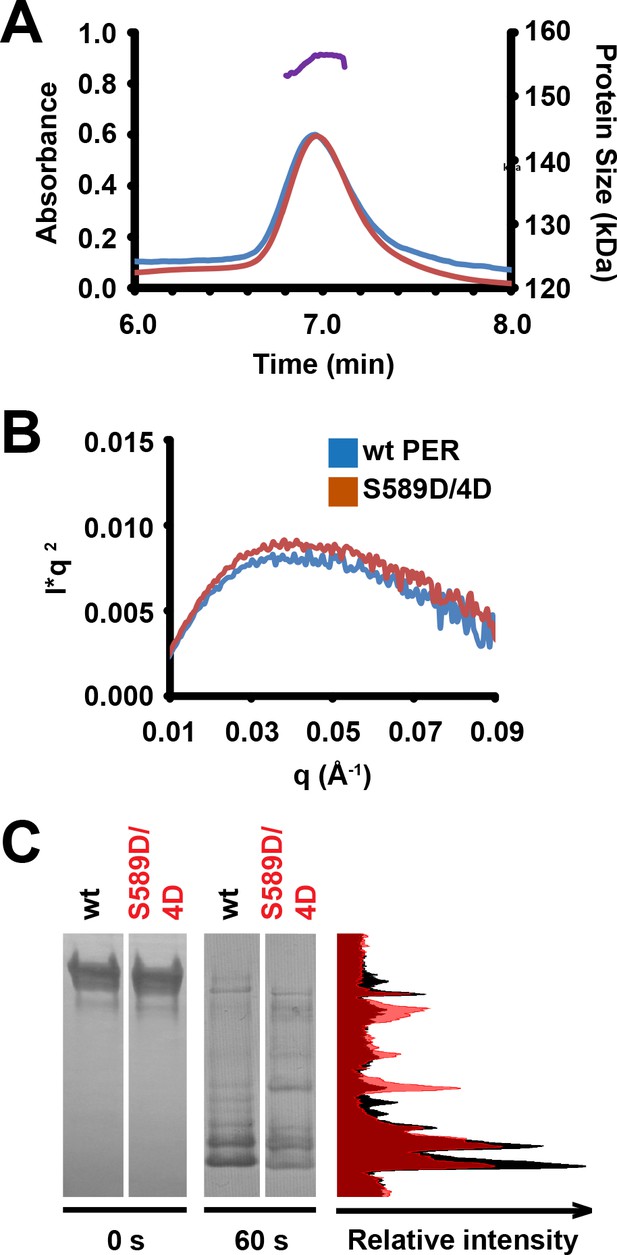
Residue substitutions of DBT target sites do not affect global conformational differences in purified PER fragment, but affect complex assembly.
(A) SEC-MALS traces of wild type (blue line) or S589D/4D (red line) PER fragments (amino acids 1–700). Absorbance is measured at 280 nm across time, peaking at ~0.6. The purple peak represents the molecular weight of the wt/mutant PER fragments, corresponding to ~155 kDa. (B) Kratky plots of wt PER fragment and the S589D/4D mutant derived from small-angle x-ray scattering (SAXS). l: scattered intensity, q: scattering vector. (C) Wildtype (wt) or mutant (S589D/4D) PER fragments (amino acids 1–700) were exposed to trypsin digestion for 0 or 60 s, and analyzed by SDS-PAGE gel and Coomassie stain. Representative image shown. Band intensities of wild-type (black curve) and mutant (red curve) PER were quantified, plotted and aligned with the bands of the SDS-PAGE gel for visual clarity of band intensities.
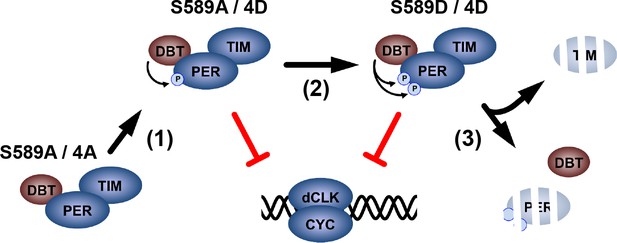
A proposed model for DBT-mediated PER regulation.
Non-phosphorylated PER is stable, but unable to inhibit dCLK-mediated transcription. DBT binds PER and phosphorylates the PER-SD to activate PER inhibition of dCLK (Step 1). DBT phosphorylates S589 stabilizing the PER/TIM complex, while PER continues to inhibit dCLK (Step 2). Subsequently, TIM is degraded, releasing PER for degradation, ending the PER life cycle.
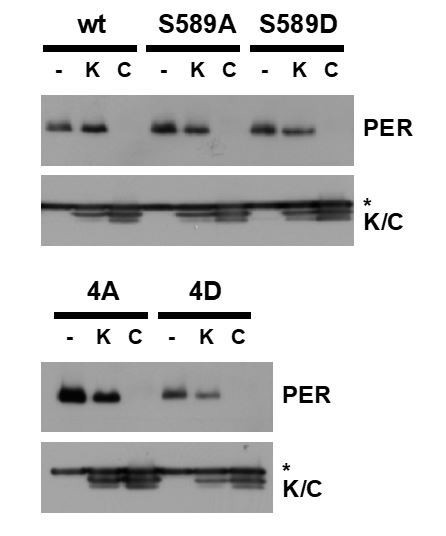
PER stability in steady state conditions in cultured cells.
Wild type PER and PER variants S589A, S589D, 4A and 4D were exogenously expressed in cultured cells, or co-expressed with wild type DBT (C) or DBT-K38R (K). Protein extracts from cells were analyzed by immunoblot using antibody against myc tag (PER) or FLAG tag (DBT). The asterisk denotes a non-specific protein that cross reacts with the FLAG antibody. These data suggest that residue substitutions in the PERS and PER-SD regions do not block DBT-mediated PER degradation. TIM was not exogenously expressed in these experiments.
Additional files
-
Supplementary file 1
Fly behavioral data.
Behavioral periods of flies charted in Figure 1B, C and D are detailed. Tau: behavioral period, SD: standard deviation, No. Arr.: Number of arrhythmic flies, Total Number: Total number of flies tested. fs: figure supplement.
- https://doi.org/10.7554/eLife.32679.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32679.017





